Abstract
In addition to their roles in desensitization and signaling of seven-membrane-spanning receptors, β-arrestins have been more recently implicated in regulating non-seven-membrane-spanning receptor pathways. By using a yeast two-hybrid screen, we identified the inhibitor of NF-κB, IκBα, as a binding partner of β-arrestin 1. Both β-arrestin 1 and 2 interact with IκBα in transfected cells as assessed by immunoprecipitation experiments. Additionally, upstream kinases known to regulate the function of IκBα, such as IκB kinase α and β and NF-κB-inducing kinase, were also shown to interact with β-arrestin. Overexpression of either β-arrestin 1 or β-arrestin 2 led to marked inhibition of NF-κB activity, as measured by reporter gene activity. Inhibition of NF-κB activity was independent of the type of stimulus used for NF-κB activation. Conversely, suppression of β-arrestin 1, but not β-arrestin 2, expression by using RNA interference led to a 3-fold increase in tumor necrosis factor-stimulated NF-κB activity as measured by NF-κB mobility-shift analysis. These data uncover a role of β-arrestins in the regulation of NF-κB-mediated gene regulation.
Arrestins are cytosolic proteins that play a critical role in the regulation of seven-membrane-spanning (7MS) receptor signaling. The arrestin family consists of four isoforms, two expressed only in the visual system (visual and cone arrestin) and two that are ubiquitously expressed, β-arrestins 1 and 2 (1–3). The classical paradigm for desensitization of 7MS receptors involves phosphorylation of the ligand-bound receptor by G protein-coupled receptor kinases, followed by the recruitment of arrestin proteins (4, 5). Arrestin binding blocks G protein coupling and mediates receptor endocytosis. More recently, β-arrestins have been shown to link 7MS receptors to the activation of other signaling pathways, such as mitogen-activated protein kinase cascades (6, 7). The ability to recruit β-arrestins and use their scaffolding properties is not limited to 7MS receptors. For instance, the insulin-like growth factor 1 receptor, a receptor tyrosine kinase, activates phosphatidylinositol 3-kinase in a β-arrestin-dependent manner (8). Thus, in addition to the traditional view of arrestins as negative regulators of 7MS signaling, they are also able to initiate signaling from a variety of receptors.
NF-κB is a ubiquitously expressed transcription factor that regulates genes involved in immune regulation, cell migration, inflammation, and apoptosis. Mammalian cells express five members of the NF-κB/Rel family: p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), c-Rel, and RelB, which form heterodimers and homodimers in the cell. In the inactive state, NF-κB dimers are retained in the cytosol because of their association with inhibitory proteins called IκBs (9). There are five mammalian IκB proteins, with IκBα and IκBβ being the best studied and understood (10). A wide variety of stimuli, such as cytokines, oxidative stress, infection, and 7MS receptor agonists (11), converge on a kinase complex consisting of NF-κB-inducing kinase (NIK) and IκB kinase (IKKα, IKKβ, and IKKγ). The catalytic components of the kinase complex (IKKα and IKKβ) phosphorylate Ser-32 and Ser-36 of IκBα (12, 13), targeting it for degradation by means of the ubiquitination and subsequent degradation by the 26S proteosome pathway (14–16). After degradation of IκBα, the nuclear targeting signal of NF-κB is unmasked, allowing it to translocate to the nucleus, where it can bind to specific κB sites to promote transcription. Activation of NF-κB can also be achieved after tyrosine phosphorylation of IκBα at Tyr-42, an event that also ultimately leads to the dissociation of NF-κB and IκBα (17, 18).
The regulation of NF-κB activity depends on its interaction with IκB. Thus, proteins that bind to IκB are critical regulators of NF-κB activity. Several proteins have been shown to interact with IκB, including the retinoic acid receptor (19), the catalytic subunit of protein kinase A (20), and κB-ras proteins (21). Here, we identify β-arrestin as a binding partner of IκBα and characterize its effect on NF-κB signaling.
Materials and Methods
Materials. Human tumor necrosis factor α (TNF-α), human angiotensin II, and carbachol were obtained from Promega, Peninsula Laboratories, and Sigma, respectively. Pervanadate was prepared fresh by adding 50 mM Na3VO4 to 50 mM H2O2. After incubating the mixture at room temperature, we added 100 units of catalase to quench the reaction. The pervanadate solution was used immediately.
Yeast Two-Hybrid Screening. A rat β-arrestin 1 cDNA (containing a single point mutation converting Arg-161 to Gly) was cloned into the pAS2-1 yeast expression vector (Clontech). The pAS2-1 (β-arrestin 1) plasmid was transformed into the PJ-69-4A yeast strain with a human heart cDNA library (Clontech) by following standard yeast transformation protocols (22, 23). Rescued library plasmids from positive clones were sequenced with an ABI DNA sequencer (Howard Hughes Nucleic Acid Facility, Duke University). The isolated plasmids were then cotransformed into yeast with either the pAS2–1 (β-arrestin 1) plasmid or pAS2–1, and the specificity of the interactions were confirmed by growth on His- and Ade-selective plates.
Plasmids. pCMV-IκBα and pGL2 3×κB-luciferase plasmids were obtained from Al Baldwin (University of North Carolina, Chapel Hill) (24, 25). pRK-FLAG IκBα, pRK-FLAG IKKα (26), pRK-FLAG IKKβ (27), and pRK-FLAG-NIK (28) were obtained from David Goeddel (Tularik, San Francisco). pCMV-β-arrestin 1 and pCMV-β-arrestin 2 were described in ref. 2. The constructs for pcDNA3-FLAG β-arrestins 1 and 2 were described in refs. 29 and 30. The pRL-TK plasmid for the Renilla luciferase control was purchased from Promega. The expression plasmids encoding hemagglutinin epitope-tagged AT1A receptor (pcDNA3.1-HA-AT1AR) and muscarinic M1 receptor (pRK5-HA-M1R) plasmids were provided by M. G. Caron (Duke University).
Cell Culture and Transfection. HeLa cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin in a 37°C incubator under 5% CO2. Jurkat T cells were maintained in RPMI medium 1640 supplemented with 10% FBS and 1% penicillin/streptomycin in a 37°C incubator under 5% CO2. Cells were transfected with the FuGENE 6 reagent (Roche Applied Sciences) by using a 3:1 ratio (μl of FuGENE:μg of DNA) according to the manufacturer's instructions.
Western Blot Analysis. Equal amounts of cell lysate (as determined by Bradford assay) were resolved on 10% Tris-glycine polyacrylamide gels (Invitrogen). After transfer to nitrocellulose, Western blot analysis was performed with the appropriate antibody. For β-arrestin detection, a 1:3,000 dilution of β-arrestin A1CT (2) was used. For detection of the FLAG epitope, anti-FLAG M2 antibody (Sigma) was used at a 1:1,000 dilution. Detection of IκBα was performed by using polyclonal IκBα antibody (C15, Santa Cruz Biotechnology) at a 1:500 dilution. Anti-mouse and anti-rabbit horseradish peroxidase-coupled secondary antibodies were purchased from Jackson Immunologicals, West Grove, PA.
Immunoprecipitation. Forty-eight hours after transfection, cells from a 10-cm plate were lysed with 1 ml of buffer containing 20 mM Tris·HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.5% Nonidet P-40, and protease inhibitors. After incubation for 30 min on ice, the samples were cleared by centrifugation at 21,000 × g for 10 min at 4°C. The supernatant fluid was added to a ≈50-μl bed volume of anti-FLAG M2 affinity gel (Sigma) that was preequilibrated with lysis buffer. The samples were immunoprecipitated with constant mixing at 4°C for at least 4 h. The beads were then washed five times with lysis buffer, transferred to a new tube, eluted with 2× SDS/PAGE sample buffer, and analyzed by SDS/PAGE and Western blot.
NF-κB Reporter Assay. HeLa or Jurkat T cells were transfected in six-well plates as described above by using FuGENE 6. For each transfection, 120 ng of the 3×κB-luciferase reporter plasmid and 20 ng of the pRH-TK control plasmid were used in a total of 1 μg of DNA. When indicated, additional receptors or β-arrestin plasmids were also included. The following day, cells were serum-starved overnight with DMEM plus 1% penicillin/streptomycin without FBS. Cells were then treated for 4–6 h, washed once with PBS, and then analyzed with the Dual-Luciferase reporter assay system (Promega), according to the manufacturer's instructions, with a Lumat LB 9507 luminometer (Berthold, Nashua, NH). The luciferase data were plotted and analyzed by using prism software (GraphPad, San Diego).
Electrophoretic Mobility-Shift Assay. HeLa cells were split in 10-cm plates to result in 50–60% confluence the following day. The cells were then transfected with control, β-arrestin 1, or β-arrestin 2 small inhibitory RNA (siRNA) oligonucleotides (31) by using the GeneSilencer reagent (Gene Therapy Systems, San Diego) according to the manufacturer's instructions. Forty-eight hours after transfection, three 10-cm plates were split, combined, and replated into 10-cm plates to have a homogenous cell population for each time point. The next day, the cells were stimulated with 10 ng/ml TNF-α for 0, 10, or 30 min. After stimulation, the cells were immediately washed once with ice-cold PBS then harvested in ice-cold PBS and placed on ice. Nuclear extracts were prepared by first pelleting the cell suspension at 1,100 × g for 5 min. The resulting pellet was resuspended in cytoplasmic extract buffer (10 mM Hepes, pH 8.0/60 mM KCl/1 mM EDTA/1 mM DTT/1 mM PMSF) with 0.3% Nonidet P-40. After a 5-min incubation, the samples were again centrifuged at 1,100 × g for 5 min. The pellet was washed with cytoplasmic extract buffer without Nonidet P-40 and pelletted once more at 1,100 × g for 5 min. The resulting pellet was resuspended in nuclear extract buffer (20 mM Tris, pH 8.0/420 mM NaCl/1.5 mM MgCl2/0.2 mM EDTA/1 mM PMSF/25% glycerol), incubated on ice for 10 min with occasional mixing, and cleared at 21,000 × g for 10 min. The resulting supernatant fluid represented the nuclear extract and was used for subsequent gel-shift analysis.
The NF-κB consensus oligonucleotide was purchased from Promega. Labeling of the probe with [γ-32P]ATP was done according to the manufacturer's instructions. Each reaction contained 2 μg of nuclear extract and 1.75 pmol of radiolabeled oligonucleotide and was carried out for 30 min at room temperature. For supershift analysis, the nuclear extracts were preincubated for 30 min with 1 μl of NF-κB p50 or p65 antibody (Upstate Biotechnology, Lake Placid, NY). The samples were loaded in the absence of loading dye onto a 6% DNA retardation gel (Invitrogen), which had been prerun for 1 h, and ran at 10 V for 75 min. After electrophoresis, the gel was dried onto filter paper, analyzed with a Typhoon 9200 PhosphorImager (Molecular Dynamics), and quantified by using imagequant 5.0 software.
Results
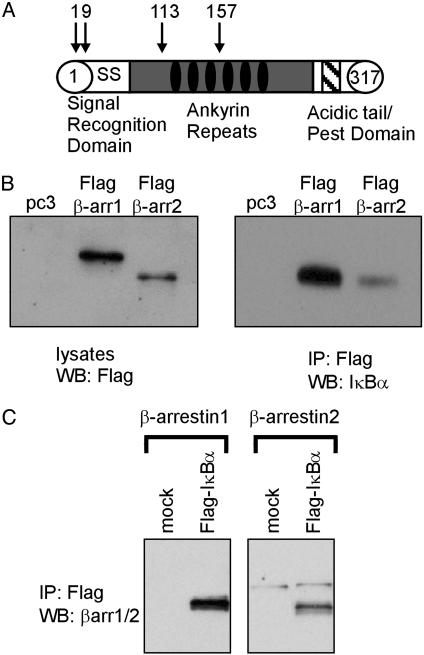
Identification of IκBα as a β-Arrestin 1 Interacting Protein. To search for β-arrestin 1 interacting proteins, we screened a human heart cDNA library by using a β-arrestin 1–GAL4 DNA binding domain fusion protein as bait in the yeast two-hybrid system. From 4 × 106 independent transformants, we obtained four cDNA clones that encode the NF-κB regulatory protein IκBα (Fig. 1A). The longest clone represented amino acids 1–317 of IκBα, whereas shorter clones encoded amino acids 9–317, 113–317, and 157–317 of IκBα. To confirm this interaction, the β-arrestin bait plasmid and the isolated cDNA clone encoding amino acids 113–317 of IκBα were cotransformed into yeast, and this combination demonstrated specific interaction when analyzed in the two-hybrid system (data not shown).
Fig. 1.
Identification of IκBα as a β-arrestin-interacting protein. (A) Yeast two-hybrid screening of a human heart cDNA library with β-arrestin 1 identified four independent clones of IκBα. The basic structure of IκBα and the positions of the 5′ end of the identified clones are depicted. (B) HeLa cells overexpressing either FLAG β-arrestin 1 (β-arr1) or 2 (β-arr2) along with IκBα were immunoprecipitated (IP) with anti-FLAG antibodies. Western blots (WB) depict total cell lysates probed with anti-FLAG antibody (Left) and FLAG-immunoprecipitates analyzed with anti-IκBα antibody (Right). (C) HeLa cells overexpressing FLAG IκBα along with either β-arrestin 1 or 2 were immunoprecipitated with anti-FLAG antibodies. Western blots are immunoprecipitates stained with anti-β-arrestin antibody. Data shown are representative of four independent experiments.
To demonstrate the interaction in mammalian cells, we transfected HeLa cells with plasmids encoding FLAG-β-arrestins and IκBα. By using antibodies against the FLAG epitope, we were able to immunoprecipitate IκBα along with FLAG β-arrestin 1 and β-arrestin 2 (Fig. 1B). The reciprocal experiment also showed that β-arrestins 1 and 2 immunoprecipitated with FLAG IκBα (Fig. 1C). These cellular data confirm the yeast two-hybrid experiment and demonstrate that both β-arrestin 1 and 2 interact with IκBα when overexpressed in HeLa cells.
β-Arrestin Interacts with the IKK Complex. β-Arrestins have been well characterized in their ability to simultaneously bind multiple signaling proteins. To determine whether β-arrestins serve a similar role in the IκB signaling pathway, we transfected HeLa cells with β-arrestins 1 and 2 along with FLAG-tagged IKKα, IKKβ, or NIK and immunoprecipitated the cell lysates with anti-FLAG beads. As shown in Fig. 2, β-arrestins 1 and 2 were coimmunoprecipitated with IKKα, IKKβ, and NIK. This result indicates that β-arrestin can also interact with the kinases involved in IκBα regulation, as well as IκBα itself.
Fig. 2.
β-arrestin (β-arr1/2) interacts with members of the IKK complex. HeLa cells were transfected with the indicated plasmids. After transfection (48 h), cells were lysed and immunoprecipitated (IP) by using anti-FLAG M2 beads. Western blot analysis was performed by using anti-β-arrestin antibody. Data shown are representative of four independent experiments.
β-Arrestin Attenuates NF-κB Activity. To examine the effect of β-arrestins on NF-κB activity, we used a transcriptional reporter assay. Overexpression of β-arrestins 1 or 2 in HeLa cells attenuated NF-κB activity stimulated by TNF-α, a prototypical NF-κB-inducing cytokine (32–34), by ≈75% (Fig. 3A). Furthermore, stimulation of cells with the muscarinic receptor agonist carbachol (in cells overexpressing the M1 muscarinic receptor; Fig. 3A) and the protein tyrosine phosphatase inhibitor pervanadate (data not shown) also led to similar levels of NF-κB inhibition.
Fig. 3.
Attenuation of NF-κB activation by β-arrestins. (A) HeLa or Jurkat T cells were transfected with an NF-κB reporter gene along with β-arrestin 1 (β-arr1, gray), β-arrestin 2 (β-arr2, white), or pcDNA3 (black) as a control. For carbachol-stimulated experiments, cells were also transfected with plasmid for the M1 muscarinic receptor. After overnight serum starving, cells were stimulated for 4 h with 10 ng/ml TNF-α or 100 μM carbachol. (B) HeLa cells were treated as in A with the exception of being transfected with FLAG β-arrestin 1 and FLAG β-arrestin 2 constructs. Levels of β-arrestin expression are shown by Western blot (WB) detection with FLAG antibody (Inset). Cells were treated with either 10 ng/ml TNF-α or 100 μM angiotensin for 4 h. Data represent the mean ± SE from three independent experiments performed in triplicate. Statistical significance was determined by using a one-way ANOVA to compare control with β-arrestin 1 and β-arrestin 2 transfected cells by using prism software. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether the inhibition by β-arrestins is a general phenomenon observed in multiple cellular backgrounds, Jurkat T cells were also analyzed. Similar to the results in HeLa cells, NF-κB activation in response to TNF-α and carbachol (in cells overexpressing the M1 muscarinic receptor) was attenuated by overexpression of either β-arrestin 1 or β-arrestin 2 (Fig. 3A). NF-κB activity stimulated by pervanadate was also attenuated in Jurkat cells (data not shown). Because our β-arrestin-specific antibody detects β-arrestin 1 more robustly than β-arrestin 2 (data not shown), we used FLAG-tagged constructs of β-arrestins 1 and 2 to determine whether either isoform was a more effective inhibitor of NF-κB activation in cells. At equal levels of expression for β-arrestins 1 and 2 (Fig. 3B Inset), no difference in NF-κB inhibition through either TNF-α or angiotensin (by using cells overexpressing the angiotensin IA receptor) was observed (Fig. 3B). These results indicate that both β-arrestin 1 and 2 are equally effective inhibitors of NF-κB activity when overexpressed in HeLa cells.
Suppression of β-Arrestin 1, but Not β-Arrestin 2, Enhances NF-κB Activity. The ability to suppress β-arrestin expression by using siRNA in mammalian cells has been recently described (31). We applied this technique to HeLa cells to determine the effect suppression of endogenous β-arrestin expression has on NF-κB activity. Our attempts to combine siRNA transfection with the reporter gene assay were unsuccessful, because the siRNA transfection reagent directly induced the NF-κB-mediated transcription (data not shown). To circumvent this problem, we used electrophoretic mobility gel-shift assays to study the effect of siRNA transfection for β-arrestins 1 and 2 on NF-κB activity. As illustrated in Fig. 4, decreasing β-arrestin 1 expression led to enhanced NF-κB activity in response to treatment with TNF-α compared with cells transfected with a control siRNA. Surprisingly, decreasing levels of β-arrestin 2 did not increase NF-κB activity. The NF-κB activity observed in HeLa cells was demonstrated to be caused by the p50/p65 heterodimer, as shown by supershift analysis (Fig. 4A). Quantification of the data (Fig. 4B) demonstrated that after a 30-min stimulation with TNF-α, suppression of β-arrestin 1 expression increased NF-κB activity by 3.4-fold compared with control, whereas suppression of β-arrestin 2 had no effect. Levels of β-arrestin 1 and β-arrestin 2 in the cell lysates were reduced by >80% in each experiment (Fig. 4C), indicating the specificity was indeed due to the function of β-arrestin 1 and not due to discrepancies in the level of reduction of the proteins. This result suggests that endogenous levels of β-arrestin 1 regulate the NF-κB response to TNF-α.
Fig. 4.
Transfection of β-arrestin 1, but not β-arrestin 2, siRNA leads to an increase in NF-κB activity. (A) HeLa cells were transfected with siRNA for β-arrestin 1(βarr1), β-arrestin 2 (βarr2), or control (CTL), treated with 10 ng/ml TNF-α, and subjected to gel-shift mobility assays as described in Materials and Methods. A representative mobility shift experiment (Left) and a supershift experiment with antibodies to the p50 and p65 subunits of NF-κB(Right) are shown. (B) The NF-κB band intensity was quantified by using a PhosphorImager. The data represent the mean ± SE (n = 7). Statistical significance was determined by using a one-way ANOVA to compare control-transfected cells to β-arrestin 1 and β-arrestin 2 siRNA-transfected cells after 30 min of stimulation. *, P < 0.05. (C) Western blot of lysates from cells transfected with control, β-arrestin 1, and β-arrestin 2 siRNAs probed with anti-β-arrestin antibody.
Discussion
Traditionally thought to serve an integral role in the desensitization of signaling through 7MS receptors, arrestin proteins have additionally been shown to serve as adaptor proteins involved in the activation of intracellular signaling pathways. We have identified IκBα as an interacting partner of β-arrestin 1 in a yeast two-hybrid screen. This interaction was confirmed in immunoprecipitation experiments, showing that both β-arrestin 1 and 2 interact with IκBα (Fig. 1). The function of β-arrestin as an inhibitor of NF-κB activity was first demonstrated by the attenuation of NF-κB activation upon overexpression of β-arrestin 1 or 2.
Based on the profound effect of β-arrestin overexpression on NF-κB activity, we then examined the effect of suppressing the expression of endogenous β-arrestins on NF-κB activity. Reduction of endogenous β-arrestin 1, but not β-arrestin 2, levels led to an increase in NF-κB activity in response to TNF-α (Fig. 4). This result suggests that at endogenous levels, β-arrestin 1 specifically inhibits NF-κB induction, despite both β-arrestin 1 and 2 functioning to inhibit when overexpressed (Fig. 3). This apparent contradiction is presumably explained by the ability of overexpressed levels of β-arrestin 2 to mimic the effect of β-arrestin 1. A similar functional disparity between the overexpression and reduction of β-arrestin has also been observed in the extracellular signal-regulated kinase (ERK) system. Overexpression of either β-arrestin 1 or 2 results in increased ERK phosphorylation and subsequent inhibition of ERK-mediated transcription because of the ability of β-arrestin to hold ERK in the cytoplasm (35). However, suppression of endogenous β-arrestin 2 nearly eliminates ERK activation, whereas reduction of β-arrestin 1 levels increases ERK activation (31). Thus, the role of β-arrestin in both the induction of NF-κB activity and ERK activity seems to display isoform specificity at endogenous levels of β-arrestin. Our finding again demonstrates the power of siRNA technology to elucidate the specific roles of β-arrestin isoforms in signaling pathways.
NF-κB can be activated by a variety of different stimuli. TNF-α leads to activation of NF-κB by the traditional mechanism of IκB degradation after phosphorylation by the IKK complex. Similarly, signaling through 7MS receptors can lead to this traditional serine phosphorylation of IκBα and subsequent activation of NF-κB (36, 37). IκB inhibition of NF-κB can also be released by means of a tyrosine phosphorylation event. Pervanadate, a protein tyrosine phosphatase inhibitor, induces IκBα tyrosine phosphorylation and activation of NF-κB (38). Overexpression of β-arrestin inhibited NF-κB activity through both serine (induced by TNF-α, carbachol, and angiotensin) and tyrosine (induced by pervanadate) phosphorylation of IκBα (Fig. 3), indicating that β-arrestin inhibited NF-κB activity independent of the mechanism of activation. One possible role of β-arrestin in this pathway may be to localize the IκB-NF-κB dimer in the cytosol. Such a role would be similar to the β-arrestin-mediated cytosolic retention of phospho-ERK induced by either angiotensin or proteinase-activated receptor 2 activation, which also prevents ERK-dependent transcription (35, 39). In addition to ERK-dependent transcription, β-arrestin 1 also modulates the transcriptional activity of the lymphoid enhancer factor (40). This apparent involvement of β-arrestin in the NF-κB signaling paradigm expands its function in regulating gene transcription.
Our data identify a pathway in which β-arrestins play a role in regulating cellular signaling. By interacting with IκBα, we demonstrate that β-arrestins attenuate NF-κB signaling. Furthermore, by using an siRNA approach, we present data that β-arrestin 1, in particular, regulates NF-κB activity, expanding recent reports of isoform-specific roles of β-arrestins. Our results further develop the role of β-arrestin as a protein responsible for the regulation of transcriptional activation. The continued study of β-arrestins will certainly lead to a more complete understanding of the importance of these proteins in regulation of gene transcription pathways.
Acknowledgments
We thank Donna Addison for excellent secretarial assistance. This work was supported by National Institutes of Health Grant RO1 HL16037 (to R.J.L.). R.J.L. is an investigator of the Howard Hughes Medical Institute.
Abbreviations: 7MS, seven-membrane spanning; IκB, inhibitor of NF-κB; IKK, IκB kinase; TNF-α, tumor necrosis factor α; siRNA, small inhibitory RNA; ERK, extracellular signal-regulated kinase; NIK, NF-κB-inducing kinase.
References
- 1.Lohse, M. J., Benovic, J. L., Codina, J., Caron, M. G. & Lefkowitz, R. J. (1990) Science 248, 1547–1550. [DOI] [PubMed] [Google Scholar]
- 2.Attramadal, H., Arriza, J. L., Aoki, C., Dawson, T. M., Codina, J., Kwatra, M. M., Snyder, S. H., Caron, M. G. & Lefkowitz, R. J. (1992) J. Biol. Chem. 267, 17882–17890. [PubMed] [Google Scholar]
- 3.Krupnick, J. G. & Benovic, J. L. (1998) Annu. Rev. Pharmacol. Toxicol. 38, 289–319. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, J., Ferguson, S. S., Barak, L. S., Aber, M. J., Giros, B., Lefkowitz, R. J. & Caron, M. G. (1997) Receptors Channels 5, 193–199. [PubMed] [Google Scholar]
- 5.Lefkowitz, R. J. (1998) J. Biol. Chem. 273, 18677–18680. [DOI] [PubMed] [Google Scholar]
- 6.Luttrell, L. M. & Lefkowitz, R. J. (2002) J. Cell Sci. 115, 455–465. [DOI] [PubMed] [Google Scholar]
- 7.Perry, S. J. & Lefkowitz, R. J. (2002) Trends Cell Biol. 12, 130–138. [DOI] [PubMed] [Google Scholar]
- 8.Povsic, T. J., Kohout, T. A. & Lefkowitz, R. J. (2003) J. Biol. Chem. 278, 51334–51339. [DOI] [PubMed] [Google Scholar]
- 9.Baeuerle, P. A. & Baltimore, D. (1988) Science 242, 540–546. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683. [DOI] [PubMed] [Google Scholar]
- 11.Ye, R. D. (2001) J. Leukocyte Biol. 70, 839–848. [PubMed] [Google Scholar]
- 12.Brown, K., Gerstberger, S., Carlson, L., Franzoso, G. & Siebenlist, U. (1995) Science 267, 1485–1488. [DOI] [PubMed] [Google Scholar]
- 13.Traenckner, E. B., Pahl, H. L., Henkel, T., Schmidt, K. N., Wilk, S. & Baeuerle, P. A. (1995) EMBO J. 14, 2876–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palombella, V. J., Rando, O. J., Goldberg, A. L. & Maniatis, T. (1994) Cell 78, 773–785. [DOI] [PubMed] [Google Scholar]
- 15.Traenckner, E. B., Wilk, S. & Baeuerle, P. A. (1994) EMBO J. 13, 5433–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Z., Hagler, J., Palombella, V. J., Melandri, F., Scherer, D., Ballard, D. & Maniatis, T. (1995) Genes Dev. 9, 1586–1597. [DOI] [PubMed] [Google Scholar]
- 17.Imbert, V., Rupec, R. A., Livolsi, A., Pahl, H. L., Traenckner, E. B., Mueller-Dieckmann, C., Farahifar, D., Rossi, B., Auberger, P., Baeuerle, P. A. & Peyron, J. F. (1996) Cell 86, 787–798. [DOI] [PubMed] [Google Scholar]
- 18.Livolsi, A., Busuttil, V., Imbert, V., Abraham, R. T. & Peyron, J. F. (2001) Eur. J. Biochem. 268, 1508–1515. [DOI] [PubMed] [Google Scholar]
- 19.Na, S. Y., Kim, H. J., Lee, S. K., Choi, H. S., Na, D. S., Lee, M. O., Chung, M., Moore, D. D. & Lee, J. W. (1998) J. Biol. Chem. 273, 3212–3215. [DOI] [PubMed] [Google Scholar]
- 20.Zhong, H., SuYang, H., Erdjument-Bromage, H., Tempst, P. & Ghosh, S. (1997) Cell 89, 413–424. [DOI] [PubMed] [Google Scholar]
- 21.Fenwick, C., Na, S. Y., Voll, R. E., Zhong, H., Im, S. Y., Lee, J. W. & Ghosh, S. (2000) Science 287, 869–873. [DOI] [PubMed] [Google Scholar]
- 22.Gietz, R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. (1995) Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- 23.James, P., Halladay, J. & Craig, E. A. (1996) Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haskill, S., Beg, A. A., Tompkins, S. M., Morris, J. S., Yurochko, A. D., Sampson-Johannes, A., Mondal, K., Ralph, P. & Baldwin, A. S., Jr. (1991) Cell 65, 1281–1289. [DOI] [PubMed] [Google Scholar]
- 25.Cheshire, J. L. & Baldwin, A. S., Jr. (1997) Mol. Cell. Biol. 17, 6746–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regnier, C. H., Song, H. Y., Gao, X., Goeddel, D. V., Cao, Z. & Rothe, M. (1997) Cell 90, 373–383. [DOI] [PubMed] [Google Scholar]
- 27.Woronicz, J. D., Gao, X., Cao, Z., Rothe, M. & Goeddel, D. V. (1997) Science 278, 866–869. [DOI] [PubMed] [Google Scholar]
- 28.Song, H. Y., Regnier, C. H., Kirschning, C. J., Goeddel, D. V. & Rothe, M. (1997) Proc. Natl. Acad. Sci. USA 94, 9792–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, F. T., Miller, W. E., Luttrell, L. M. & Lefkowitz, R. J. (1999) J. Biol. Chem. 274, 15971–15974. [DOI] [PubMed] [Google Scholar]
- 30.Miller, W. E., Maudsley, S., Ahn, S., Khan, K. D., Luttrell, L. M. & Lefkowitz, R. J. (2000) J. Biol. Chem. 275, 11312–11319. [DOI] [PubMed] [Google Scholar]
- 31.Ahn, S., Nelson, C. D., Garrison, T. R., Miller, W. E. & Lefkowitz, R. J. (2003) Proc. Natl. Acad. Sci. USA 100, 1740–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osborn, L., Kunkel, S. & Nabel, G. J. (1989) Proc. Natl. Acad. Sci. USA 86, 2336–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowenthal, J. W., Ballard, D. W., Bogerd, H., Bohnlein, E. & Greene, W. C. (1989) J. Immunol. 142, 3121–3128. [PubMed] [Google Scholar]
- 34.Duh, E. J., Maury, W. J., Folks, T. M., Fauci, A. S. & Rabson, A. B. (1989) Proc. Natl. Acad. Sci. USA 86, 5974–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tohgo, A., Pierce, K. L., Choy, E. W., Lefkowitz, R. J. & Luttrell, L. M. (2002) J. Biol. Chem. 277, 9429–9436. [DOI] [PubMed] [Google Scholar]
- 36.Todisco, A., Ramamoorthy, S., Pausawasdi, N. & Tacey, K. (1999) Biochem. Biophys. Res. Commun. 261, 877–884. [DOI] [PubMed] [Google Scholar]
- 37.Purcell, N. H., Tang, G., Yu, C., Mercurio, F., DiDonato, J. A. & Lin, A. (2001) Proc. Natl. Acad. Sci. USA 98, 6668–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, S. & Aggarwal, B. B. (1995) J. Biol. Chem. 270, 10631–10639. [DOI] [PubMed] [Google Scholar]
- 39.DeFea, K. A., Zalevsky, J., Thoma, M. S., Dery, O., Mullins, R. D. & Bunnett, N. W. (2000) J. Cell Biol. 148, 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, W., Hu, L. A., Semenov, M. V., Yanagawa, S., Kikuchi, A., Lefkowitz, R. J. & Miller, W. E. (2001) Proc. Natl. Acad. Sci. USA 98, 14889–14894. [DOI] [PMC free article] [PubMed] [Google Scholar]