Background: Both autophagy and the heat stress response represent protein management alternatives for the stressed cell. Their inter-relationship is not known.
Results: Heat shock factor-1 knockdown increases and HSP70 overexpression inhibits autophagy in cell culture model. Preactivation of heat shock inhibits exercise-induced autophagy in humans.
Conclusion: Heat shock response plays a negative role in autophagy regulation.
Significance: Heat shock response regulates autophagy.
Keywords: Akt, Autophagy, Exercise, Heat Shock Protein, mTOR, Protein Folding
Abstract
The eukaryotic cell depends on multitiered homeostatic systems ensuring maintenance of proteostasis, organellar integrity, function and turnover, and overall cellular viability. At the two opposite ends of the homeostatic system spectrum are heat shock response and autophagy. Here, we tested whether there are interactions between these homeostatic systems, one universally operational in all prokaryotic and eukaryotic cells, and the other one (autophagy) is limited to eukaryotes. We found that heat shock response regulates autophagy. The interaction between the two systems was demonstrated by testing the role of HSF-1, the central regulator of heat shock gene expression. Knockdown of HSF-1 increased the LC3 lipidation associated with formation of autophagosomal organelles, whereas depletion of HSF-1 potentiated both starvation- and rapamycin-induced autophagy. HSP70 expression but not expression of its ATPase mutant inhibited starvation or rapamycin-induced autophagy. We also show that exercise induces autophagy in humans. As predicted by our in vitro studies, glutamine supplementation as a conditioning stimulus prior to exercise significantly increased HSP70 protein expression and prevented the expected exercise induction of autophagy. Our data demonstrate for the first time that heat shock response, from the top of its regulatory cascade (HSF-1) down to the execution stages delivered by HSP70, controls autophagy thus connecting and coordinating the two extreme ends of the homeostatic systems in the eukaryotic cell.
Introduction
Protein management is an evolutionary early and highly conserved cellular function. Both prokaryotes and eukaryotes rely on the heat shock protein (HSP)2 chaperone system to fold and assemble proteins, as well as to refold or facilitate the breakdown of irreversibly denatured proteins resulting from stress. Eukaryotes additionally employ autophagy (1, 2) to sequester and deliver for degradation to lysosomes large protein aggregates and whole damaged organelles inaccessible to smaller proteolytic systems in the cell. Thus, the heat shock response and macroautophagy represent two ends of the spectrum of cellular protein quality control with the former being ubiquitous in all living organisms, whereas the latter is restricted to eukaryotic cells. Under conditions of cellular stress, these two systems are likely to complement each other. Surprisingly, their interactions and coordination of function have not been investigated. Given the significant evolutionary distances between these protein management systems, and the fact that one (HSP) has a protein conservation function, we hypothesized that cells may prioritize the HSP response over autophagy and that the HSP system would exert a control over autophagy under conditions where both could be activated.
Autophagy can be divided into several main steps (3). Initially, a portion of the cytoplasm or a damaged organelle is engulfed by double- or multimembrane autophagic vacuoles or autophagosomes, followed by completion of the autophagosomes that finally fuse with lysosomes to form autolysosomes leading to degradation of the captured material (3, 4). At basal levels, autophagy is detected in all eukaryotic cells clearing misfolded and unfolded proteins. Under starvation, autophagy represents a physiological response providing biofuel from degraded macromolecules to maintain sufficient ATP production for adaptive macromolecular synthesis to survive stressful conditions (5, 6).
Autophagy is a highly controlled and orchestrated process (3, 4). Among numerous autophagy-related proteins (ATG proteins) and kinases, serine/threonine kinase mammalian target of rapamycin (mTOR) is considered a critical negative regulator in autophagy induction. mTOR activity is regulated upstream by several kinases. Akt activates mTOR and inhibits autophagy through activation of the mTOR upstream regulator tuberous sclerosis complex 1/2 (TSC1/2) (7). Similarly, the IκB kinase complex (8), ERK1/2 pathway (9–11), and adenosine monophosphate-activated protein kinase signaling (12) are positive regulators of the autophagy induction via mTOR inhibition. Interestingly, p53 plays a dual role in autophagy regulation. On the one hand, p53 activation potentiated autophagy through inhibition of the mTOR pathway (13). On the other hand, induction of autophagy was initiated by utilizing knock-out, knockdown, or pharmacological inhibition of p53 (14).
Exposure to a variety of stresses, including heat, radiation, or heavy metals, can lead to protein denaturation, damage of nucleic acid, and even death. In addition to external stressors, homeostasis of the organism may often be compromised by internal physiological and pathological inducers, including exercise, ischemia, and inflammation. To survive this ever-changing environment, single cells and organisms have the ability to cope with the stress-induced damage by up-regulating the specific proteins called heat shock proteins. HSP70 offers protection against the damaging factors at the level of macromolecules, single cells, as well as whole organisms. HSPs, as molecular chaperones, assist in correct folding of newly synthesized proteins, translocation, degradation, and reactivation of damaged proteins (15–19). Cells exposed to acute thermal stress associated with overexpression of HSPs survived the potentially lethal effects of subsequent exposure to heat (20), serum-free medium-induced apoptosis (21), oxidative stress (22), and heavy metals (23). Exposure to heat stress protected animals to otherwise lethal effects of endotoxin (24–26). In Madin-Darby canine kidney epithelial cells, heat stress preconditioning was associated with increased maintenance of epithelial barrier function in response to sever heat exposure (15, 27), and this effect was discussed to be related to an increase in occludin tight junction protein expression (28, 29). Recently, our group has demonstrated that HSP70 overexpression regulated cytokine levels in animals (30) in a manner similar to heat conditioning (31, 32).
There are several observations showing that cellular chaperone may also play a regulatory role in autophagy. The chemically induced endoplasmic reticulum (ER) stress resulted in an increase in autophagy leading to cell death. This effect was partially associated with down-regulation of the Akt/TSC/mTOR pathway (33). A chemical chaperone, 4-phenylbutyric acid, prevented this ER-induced decrease in mTOR pathway leading to an autophagy inhibition, suggesting that cellular chaperone is important in autophagy regulation. In other studies, exposure to OSU-03012, a derivative of the cyclooxygenase-2 inhibitor celecoxib in HCT116 (human colorectal carcinoma cell line) and U251 (human glioma cell line), resulted in cell death and induction of autophagy. Transient knockdown of HSP70 expression potentiated and overexpression prevented OSU-03012-induced increase in cytotoxicity and autophagy (34). In rats, heat stress exposure (core temperature at 41 °C for 0.5 h) resulted in a significant increase in autophagy associated with subcellular injury and damage of hepatocytes, suggesting that HSPs may play a regulatory role in autophagy. Recently, it has been published that exercise induced autophagy in mice (35, 36). However, the role of HSP70 in starvation-induced autophagy still remains known. Here, we tested this model and showed that HSF-1, the central regulator of heat shock response, inhibits autophagy. The negative regulatory effect of HSF-1 on autophagy was exerted via one its principal effectors, HSP70. We also show that exercise induces autophagy in human PBMCs, and this increase in autophagy is prevented by glutamine supplementation, associated with a significant augmentation in HSP70. These data provide the first evidence directly linking the HSP and autophagy systems and demonstrate a master regulatory role of HSP70 in controlling autophagy.
EXPERIMENTAL PROCEDURES
Cells
A549 cells (human lung epithelial carcinoma cell line), Caco-2 cells (colorectal adenocarcinoma), and NCI-H1299 cells (human non-small cell lung cancer; ATCC catalog no. CRL-5803TM) were purchased from the American Type Culture Collection (Manassas, VA) and maintained at 37 °C in a culture medium composed of DMEM with 4.5 mg/ml glucose, 50 units/ml penicillin, 50 units/ml streptomycin, 4 mm glutamine, and 25 mm HEPES and supplemented with heat-inactivated 10% fetal calf serum that was purchased from Invitrogen. Stock cultures were maintained in 100-mm dishes and were subcultured every 3–4 days. Cultures for all assays were maintained in 35-mm tissue culture plates (Corning Inc., Corning, NY).
Chemicals
Rapamycin was purchased from LC Laboratories (Woburn, MA). LC3 and β-actin antibodies were purchased from Sigma. HSP70 and HSF1 antibodies were obtained from StressGen Biotechnologies (Victoria, British Columbia, Canada). CHOP and eIF2α (total and phosphor) antibodies were purchased from Cell Signaling Technology (Danvers, MA). XBP-1 antibody was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). Triton X-100, bovine serum albumin, normal donkey serum, and anti-β-actin antibody were purchased from Sigma. Horseradish peroxidase (HRP)-conjugated secondary antibodies for Western blot analysis were purchased from Cell Signaling Technology (Danvers, MA). Cy-5 antibodies for immunostaining were purchased from Jackson ImmunoResearch (West Grove, PA). Tween 20 and nonfat dry milk were purchased from Bio-Rad. All other chemicals were of reagent grade and were purchased from Sigma, VWR (West Chester, PA), or Fisher.
HSP70 Overexpression
As described previously (30), the HSP70 coding region was amplified from pRc/RSV-HSP72 provided by Dr. Anne Knowlton (University of California, Davis) using primer sets that were designed to add HindIII and XbaI sites to the 5′ and 3′ ends, respectively, and inserted into the pRc/CMV vector to construct a vector termed RP70. In a similar manner, the coding region was amplified using primers to add NotI and XbaI sites and cloned into pAdTrack (Ad70). Recombination with the adenovirus backbone was performed in BJ5183-AD-1 cells (Stratagene, La Jolla, CA), and the resulting construct was transfected into 293 cells to produce virus directing the expression of exogenous HSP70. Recombination with the parent pAdTrack (AdTrack) plasmid created the construct used as a control in all experiments. Cells were infected either with control adenovirus or adenovirus directing the expression of HSP70 (a total dose of 2.5 × 1010 IFUs per plate) for 72 h.
Site-directed Mutagenesis of HSP70
Site-directed mutagenesis of HSP70 was performed according to the to the manufacturer's instruction manual (Agilent Technologies, Inc. Santa Clara, CA).
siRNA of HSF-1
As described previously (29), to silence HSF-1, ON-TARGETplus SMARTpool (Dharmacon, Inc., Chicago) was used. The sequences for HSF-1 small interfering RNA (siRNA) were as follows: 5′-PUACUUGGGCAUGGAAUGUGUU-3′; 5′-PGUCCAUAGCAUCCAAGUGGUU-3′; 5′-PUAUGUCUUCACUCUUCAGGUU-3′; and 5′-PUGAAUCCGGGCUGCUGUUCUU-3′. A549 cells monolayers were transiently transfected using DharmaFect transfection reagent (Dharmacon, Lafayette, CO). A549 cells were seeded into a six-well plate and grown to 80% confluency. A549 monolayers were then washed with PBS, and Opti-MEM medium was added to the well. The plasmid vector containing the siRNA of HSF-1 and DharmaFect reagent was preincubated in Opti-MEM. After 5 min of incubation, two solutions were mixed and incubated for another 20 min, and the mixture was added to each well. After incubation for 3 h at 37 °C, 500 μl of DMEM containing 10% FBS and no antibiotics were added to cell culture media to reach a 2.5% final concentration of FBS. The siRNA-induced silencing of HSF-1 was confirmed by immunoblot of HSF-1.
Quantitative RT-PCR
The total RNA was isolated from A549 cells using RNeasy kit (Qiagen), and cDNA was generated using QuantiTect reverse transcription kit (Qiagen) as per the manufacturer's instructions. The quantitative RT-PCR was performed using SYBR Green I QuantiFast SYBR Green kit (Qiagen) as per the manufacturer's instructions using the following amplification conditions: PCR initial activation step, 95 °C for 5 min; two-step cycling, 40 cycles of denaturation, 95 °C for 10 s; combined annealing/extension, 60 °C for 10 s. The primers for p62 were as follows: forward primer, 5′-cacctgtctgagggcttctc-3′; reverse primer, 5′-agtttcctggtggacccatt-3′. The results were analyzed using relative quantification by comparing ratios of p62 gene and reference housekeeping gene, β-actin (internal control gene). Relative quantification of expression of the p62 gene was studied, and the fold change in expression of p62 gene was calculated relative to β-actin. The mean fold change in expression of the p62 gene was calculated using 2−ΔΔCT method.
Western Blot Analysis
For LC3 and HSF-1 activation determined by Western blot analysis and binding assay, cells were harvested and nuclear extracts were prepared. To study the effect of HSP70 protein overexpression on macroautophagy, A549, Caco-2, or NCI-H1299 monolayers were infected with the adenovirus (a total dose of 2.5 × 1010 IFUs per plate) for 3 days followed by starvation in Earle's balanced salt solution (EBSS) for 2 h. Western blot analysis was performed as described previously (28); at the end of the experimental period, monolayers were immediately rinsed with ice-cold PBS, and cells were lysed with lysis buffer (50 mm Tris·HCl, pH 7.5, 150 mm NaCl, 500 μm NaF, 2 mm EDTA, 100 μm vanadate, 100 μm PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 40 mm para-nitrophenyl phosphate, 1 μg/ml aprotinin, and 1% Triton X-100) and scraped, and the cell lysates were placed in microcentrifuge tubes. Cell lysates were centrifuged to yield a clear lysate. Supernatant was collected, and protein measurement was performed using the protein assay kit from Bio-Rad. Laemmli gel loading buffer was added to the lysate containing 5–10 μg of protein and boiled for 7 min, after which proteins were separated on an SDS-polyacrylamide gel. Proteins from the gel were transferred to the membrane (Trans-Blot Transfer Medium, Nitrocellulose Membrane; Bio-Rad) overnight. The membrane was incubated for 2 h in blocking solution (5% dry milk in TBS/Tween 20 buffer). The membrane was incubated with appropriate primary antibodies in blocking solution. After being washed in TBS/Tween buffer, the membrane was incubated in appropriate secondary antibodies and developed using the Santa Cruz Western blotting luminol reagents (Santa Cruz Biotechnology, Santa Cruz, CA) on the Kodak BioMax MS film (Fisher).
Quantitative p62 Puncta Analysis
Endogenous p62 puncta were detected by indirect immunofluorescence using a mouse anti-p62 antibody (BD Biosciences) that was detected by an AlexaFluor568-conjugated anti-mouse secondary antibody (Invitrogen). Stained samples were imaged using Cellomics Array Scan (Thermo Scientific, Rockford, IL) in which 49 fields were collected per well in 96-well plates. The mean number of p62 puncta per infected (GFP-positive) cell was then determined by a puncta-counting application integrated within the iDev software (Thermo Scientific).
Immunostaining of LC3 and HSP70 Proteins
For immunostaining of LC3 and HSP70 proteins, monolayers grown on coverslips were exposed to appropriate experimental conditions. As described previously (29), at the end of the experimental period, cells were washed twice in cold PBS and were fixed with 2% paraformaldehyde for 20 min. Then cells were permeabilized with 0.1% Triton X-100 in PBS at room temperature for 20 min. The monolayers were then incubated in blocking solution composed of bovine serum albumin and normal donkey serum in PBS for 1 h. Cells were then labeled with primary antibodies in blocking solution overnight at 4 °C. After being washed with PBS, the coverslips were incubated in appropriate Cy-5-conjugated secondary antibody for 1 h at room temperature and mounted on microscope slides (Erie Scientific, Portsmouth, NH). Immunolocalizations of TJ proteins were visualized using a Nikon fluorescence microscope (Nikon, Garden City, NY) equipped with a Hamamatsu digital camera (Hamamatsu Photonics, Hamamatsu, Japan). Images were processed with Wasabi software (Hamamatsu Photonics Deutschland, Herrsching, Germany).
Induction of Thermal Stress
To induce thermal stress, cells were grown on 35-mm plates and placed in a bath in a manner that the dishes rested directly on water that was heated to 42 °C. To maintain 5% CO2 concentration during heat exposure, air containing CO2 was circulated through the sealed bath.
Exercise Protocol and Glutamine Supplementation
Eight total endurance-trained adult men and women (age 18–45) who suffer from exercise-induced gastrointestinal distress were recruited from the University population and local running clubs. All subjects completed a health questionnaire, and a full explanation of procedures, discomforts, and risks were explained before written informed consent was obtained. Selection criteria included a report of gastrointestinal symptoms on at least five occasions during exercise and currently participating in an endurance run training program. Potential subjects were excluded if taking medications (antidepressants) or nutritional supplements. All testing was performed in the exercise laboratory at the University of New Mexico at 1524 m of altitude.
Each subject participated in a double blinded glutamine (Gln) and placebo trial, separated by a 4-week washout period. Subjects ingested 0.3 g/kg of fat-free mass, three times per day for 7 days of Gln or sugar-free lemon flavored placebo. The powder was separated into three doses that were taken in the morning, early afternoon, and evening. Supplements were mixed with water prior to drinking. Prior to the first exercise trial, subjects were provided a 7-day sample bag containing Gln or placebo. Instructions were provided detailing the proper way to take the supplement. At the end of the 7 days of supplementation, subjects performed the exercise trial.
Base-line measurements (maximal oxygen consumption, body composition, blood analysis, and gut permeability) were performed on each subject prior to the start of the study. On the day of the first exercise trial, subjects reported to the lab after an overnight fast. Height and weight were measured along with insertion of a rectal probe for core temperature measurement. The exercise was a 60-min treadmill run at 70–80% of VO2 max in a warm environmental chamber (30 °C) and with a termination criterion of 40 °C. Venous blood was collected at four time points during each exercise trial, including rest, immediately post exercise, 2 h post exercise, and 4 h post exercise.
For base-line measurements, peak aerobic power capacity (VO2 peak) was measured using open circuit spirometry (Parvomedics, Sandy, UT) and a graded incremental protocol on a motorized treadmill. The highest value for oxygen consumption based on an 11-breath running average was used to assess VO2 peak. Additional criteria, including plateau (<150 ml), respiratory exchange ratio (>1.15), rating of perceived exertion (>17), and heart rate (±10 beats/min of max), were assessed to determine whether a true maximal effort was given. Skinfold calipers were used to measured body density by calculating the sum of three sites (men: chest, abdomen, thigh; women: triceps, suprailliac, thigh), and fat-free mass was calculated using the Siri equation. After 20 min of seated rest, a 20-ml blood sample was taken to measure base-line plasma levels of glutamine, along with PBMC levels of LC3 and HSP70.
Statistical Analysis
Results are expressed as means ± S.E. Statistical significance of differences between mean values was assessed with Student's t tests for unpaired data. All reported significance levels represent two-tailed p values. A p value of <0.05 was used to indicate statistical significance.
RESULTS
Effect of HSP70 Protein Expression on Starvation-induced Autophagy
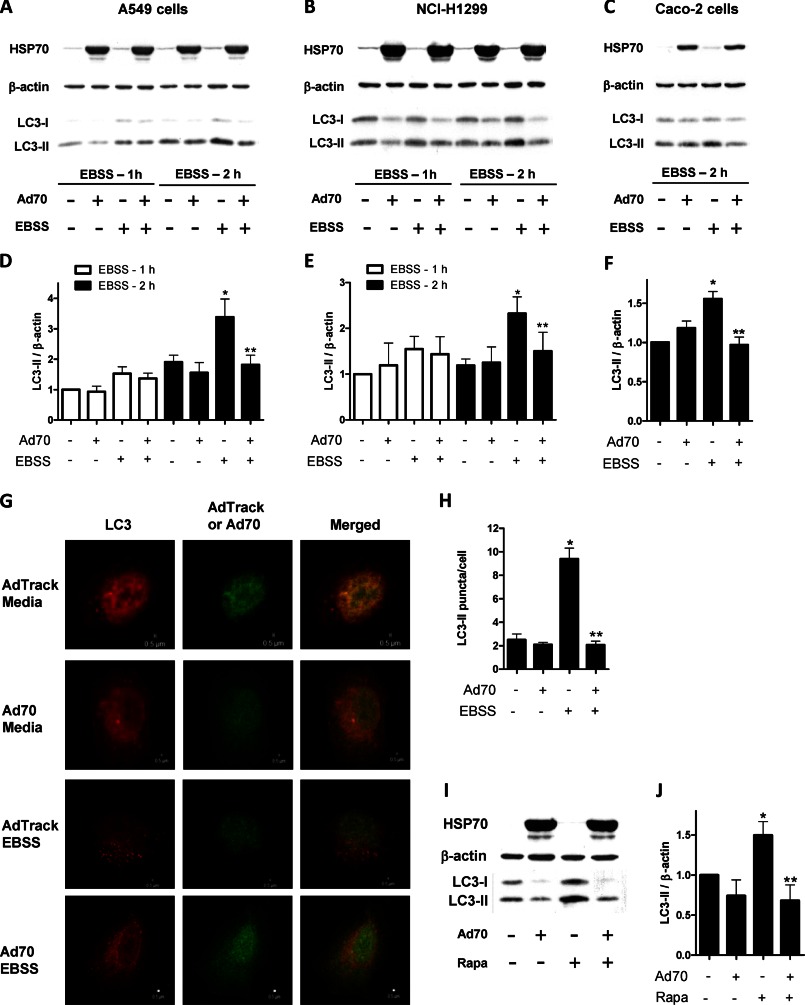
Microtubule-associated protein light chain 3 (LC3) immunoblotting was widely used to monitor the autophagy. Autophagy is manifested by conversion of the unconjugated cytosolic form of LC3-I to the phosphatidylethanolamine-conjugated form of LC3-II that targets to the autophagosomal membrane (37, 38). In the following studies, A549, H1299, or Caco-2 cells were infected with either control adenovirus or adenovirus directing expression of HSP70 (2.5 × 1010 IFUs per plate). Three days later, starvation was performed by adding EBSS to the cells along with bafilomycin A1 (Baf) (100 nm) for 1 or 2 h (39). Full medium with Baf (100 nm) was added to cells under control conditions. Cells were harvested and lysed, and aliquots were used to determine HSP70 and LC-3. The LC-3 densitometric values were normalized to β-actin protein expression as a commonly used procedure (40, 41). There was a marked increase in HSP70 protein expression in the cells infected with Ad70 compared with the cells infected with a control adenovirus (Fig. 1). At 1 h of starvation, in A549 cells there was a small and insignificant increase in LC3-II protein expression (Fig. 1, A and D). Two hours after starvation, there was a significant increase in LC3-II levels indicating an increase in autophagy. The magnitude of change is consistent with the routinely reported experimental values in the autophagy literature and summarized by Mizushima et al. (41) and Klionsky et al. (40). Interestingly, HSP70 overexpression prevented this starvation-induced autophagy, suggesting that HSP70 plays a regulatory role in modulation of autophagy. Similar effect of HSP70 on autophagy was seen in H1299 and Caco-2 cells (Fig. 1, B, C, E, and F). During autophagy induction, phosphatidylethanolamine-conjugated form of LC3 targets autophagosomal membrane resulting in the formation of punctate organelles that can be quantified by fluorescence microscopy (37). To assess the effect of HSP70 on LC3 puncta formation, A549 cells grown on coverslips were transfected with RFP-LC3. After 1 h, GFP-adenovirus (either control or overexpressing HSP70) was added to the experimental media. Following a 3-day incubation, cells were exposed to starvation conditions (EBSS with 100 nm Baf for 2 h) to induce autophagy. Cells were immunostained for puncta formation. Upon starvation, cells expressing GFP-LC3 showed a transition from diffusive cytoplasm pattern to the punctated membrane pattern, indicating the formation of autophagic vacuoles (Fig. 1, G and H). Treatment of cells with adenovirus overexpressing HSP70 decreased the number of puncta, suggesting a suppressed formation of LC3 and the translocation to the autophagosomes. In the following experiments, the effect of HSP70 overexpression on rapamycin-induced autophagy was studied (Fig. 1, I and J). Treatment of A549 cells with the mTOR inhibitor rapamycin (50 ng/ml) (39) resulted in an increased expression of LC3-II protein, and overexpression of HSP70 inhibited this up-regulation of LC3-II, indicating that HSP70 inhibits autophagy induced by rapamycin. Although in some experiments HSP70 overexpression seemed to affect the levels of the LC3-I band, the nature of the phenomenon has not been the focus of this study. We then tested whether rapamycin under control or starvation conditions affects HSP70 protein expression. To test this, A549 cells were treated with rapamycin (50 ng/ml) under control or starvation conditions (EBSS for 2 h). We found that neither rapamycin alone nor rapamycin under starvation conditions influenced HSP70 protein expression (supplemental Fig. S1).
FIGURE 1.
Effect of HSP70 overexpression on starvation- and rapamycin-induced autophagy. A549 (A and D), NCI-H1299 (B and E), and Caco2 (C and F) cells were infected with 2.5 × 1010 IFUs of either the control AdTrack or Ad70 that directs the expression of human HSP70. After a 3-day incubation, cell were exposed to either control conditions (full media with 100 nm Baf) or starvation conditions (EBSS with 100 nm Baf for 1 or 2 h), and cells were harvested and prepared for Western blot analysis to measure the relative expression of LC3, HSP70, and β-actin (an internal loading control). Densitometric values (D–F) of the corresponding blots (A–C, respectively) were obtained using Photoshop software and normalized to β-actin and set to 1 for control conditions (AdTrack virus and control culture media). D, data represent means ± S.E. (n = 3–5). *, p < 0.05 versus AdTrack/media. **, p < 0.05 versus AdTrack/EBSS. E, data represent means ± S.E. (n = 3). *, p < 0.05 versus AdTrack/media at the 2-h time point. **, p < 0.05 versus AdTrack/EBSS at the 2-h time point. F, data represents means ± S.E. (n = 3). *, p < 0.05 versus AdTrack/media at the 2-h time point. **, p < 0.05 versus AdTrack/EBSS at the 2-h time point. G, A549 cells were grown on glass coverslips and were infected with 2.5 × 1010 IFUs of either the control AdTrack or Ad70. After a 3-day incubation, cells were exposed to either control (full media with 100 nm Baf) or starvation conditions (EBSS with 100 nm Baf for 2 h). Cells were fixed and stained for HSP70 and LC3 protein expression. H, represents LC3 puncta quantification based on G; *, p < 0.05 versus AdTrack/media. **, p < 0.05 versus AdTrack/EBSS. I, A549 cells were infected with 2.5 × 1010 IFUs of either the control AdTrack or Ad70 for 3 days followed by exposure to control conditions (DMSO with 100 nm Baf) or rapamycin (50 ng/ml) with 100 nm Baf for 2 h. Cells were harvested, lysed, and prepared for Western blot analysis. J, represents LC3 autoradiogram quantification based in I. Data represent means ± S.E. (n = 3). *, p < 0.05 versus AdTrack/media. **, p < 0.05 versus AdTrack/Rapa.
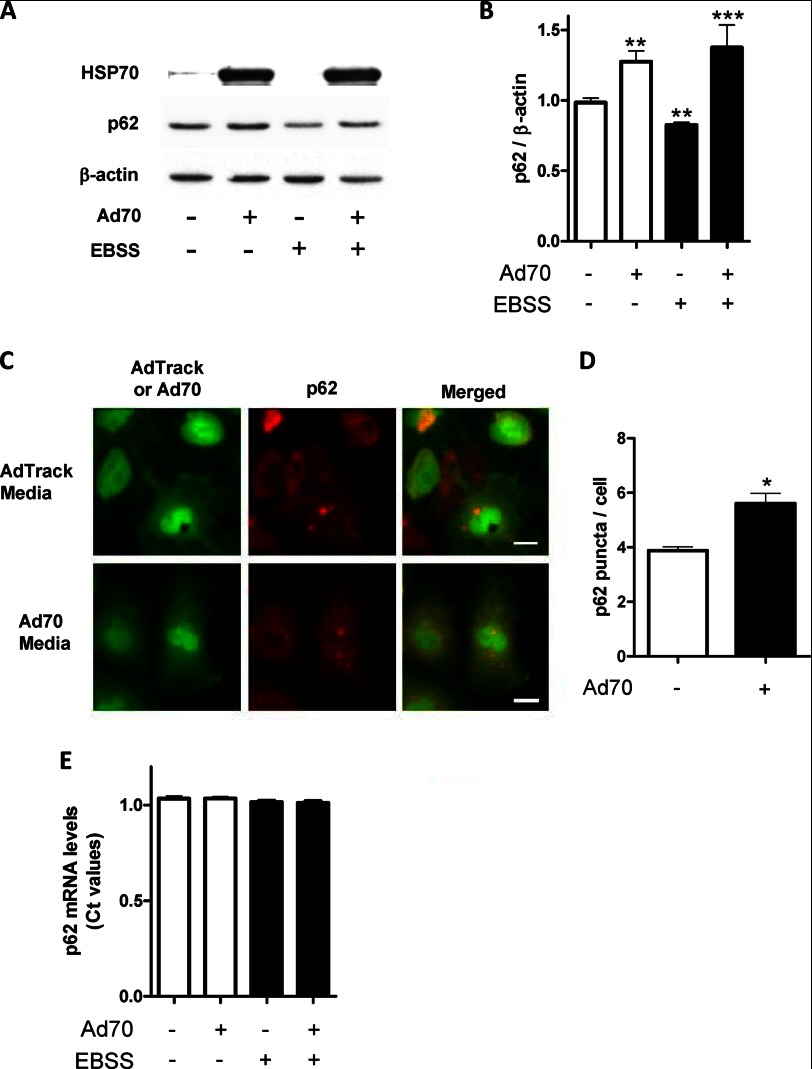
p62 is a ubiquitously expressed cellular protein that serves as a common adapter for autophagy substrates and delivers them to autophagosomes where p62 along with the substrates is degraded (42–44). In the following experiments, we tested whether HSP70 will prevent p62 degradation. A549 cells were infected with either control (AdTrack) or HSP70 overexpressing virus for 72 h. Cells were either harvested, lysed, and the autophagy substrate protein p62 determined by Western blot analysis (Fig. 2, A and B) or immunostained, and p62 formation was imaged using Cellomics Array Scan (Fig. 2, C and D). Under full media conditions, overexpression of HSP70 resulted in both a significant increase in p62 protein expression determined by Western blot analysis and p62 formation determined by immunostaining. Starvation resulted in degradation of p62, and HSP70 overexpression significantly prevented this p62 degradation. These data along with the LC3 protein expression results (Fig. 1) demonstrated that HSP70 regulates autophagy in a cell culture model. To further investigate the intracellular mechanisms involved, the effect of starvation and HSP70 interaction on p62 mRNA levels was also examined (Fig. 2E). p62 mRNA expression did not show significant change, indicating that p62 is not regulated at the mRNA level under starvation and HSP70 overexpression conditions, and the observed changes in p62 represent post-translational modifications.
FIGURE 2.
Effect of HSP70 overexpression on p62 protein expression and p62 puncta formation. A549 cells were infected with 2.5 × 1010 IFUs of either the control AdTrack or Ad70 that directs the expression of human HSP70. After a 3-day incubation, cells were harvested and prepared for Western blot analysis to measure the relative expression of p62, HSP70, β-actin (an internal loading control) (A) or p62 immunostaining (C). Densitometric values (B) of corresponding blots (A) were obtained using Photoshop software and normalized to β-actin and set to 1 for control conditions (AdTrack virus). Data represent means ± S.E. (n = 6). **, p < 0.05 versus AdTrack/media. ***, p < 0.01 versus AdTrack/EBSS. D represents p62 puncta quantification using Cellomics Array Scan based on C; *, p < 0.01 versus AdTrack. Scale bar, 20 μm. E, effect of HSP70 overexpression on mRNA p62 levels in A549 cells.
Effect of HSF-1 and High Temperature on Autophagy Regulation
The nuclear transcription factor HSF-1 has been shown to play an important role in regulating the expression of heat shock protein (45, 46). In the following experiment, we tested the hypothesis that HSF-1 plays an important role in autophagy regulation. HSF-1 expression was selectively knocked down by HSF-1 siRNA in A549 cells, and its role on autophagy regulation was studied. As shown in Fig. 3A, transfection of A549 cells with HSF-1 siRNA resulted in a marked depletion of HSF-1 expression. Under control conditions (full media with 100 nm Baf), siRNA depletion of HSF-1 resulted in a marked increase in autophagy, and HSF-1 knockdown potentiated starvation (EBSS with 100 nm Baf)-induced autophagy as demonstrated by an increase in LC3-II protein expression. These results suggest an important role of HSF-1 in regulating autophagy under both control and starvation conditions. In the following experiments, the effect of heat stress on starvation-induced autophagy was tested. A549 cells were initially exposed to heat exposure (42 °C for 2 h) followed by incubation at 37 °C for 24 h. Half of the cells were exposed to EBSS to induce autophagy under the presence of Baf (100 nm), and the other half was kept in full medium with Baf (100 nm). Cells were harvested, and HSP70, LC-3, and β-actin protein expression was determined by Western blot analysis. Heat stress exposure resulted in a significant increase in HSP70 in A549 cells (Fig. 3C). Interestingly, heat exposure resulted in a small but significant increase in autophagy as indicated by the increase in LC3-II protein expression (Fig. 3, C and D). Heat pre-conditioning in A549 cells exposed to EBSS resulted in an even larger increase in LC3-II protein expression, indicating that heat pre-conditioning potentiates starvation-induced autophagy. Next, the effect of HSF-1 on acute heat stress-induced autophagy in A549 cells was studied. First, A549 cells were transfected with HSF-1 siRNA for 48 h, followed by heat stress exposure (42 °C) for 2 h under Baf conditions. Immediately after acute heat exposure, cells were harvested, and Western blot analysis was performed. Exposure of A549 cells to HSF-1 siRNA under control temperature conditions (37 °C) led to an increase in autophagy (Fig. 3, E and F). Acute heat exposure did not result in an increase in autophagy. Cells exposed to the acute heat under HSF-1 inhibition resulted in a small autophagy inhibition compared with the HSF1 siRNA-treated cells and control temperature conditions. We also tested whether siRNA depletion of HSF-1 affected the unfolded protein response by measuring its indicators, including CHOP, XBP-1, and eIF2α (47). HSF-1 knockdown did not influence the unfolded protein response (supplemental Fig. S2). As a positive control, we utilized two commonly used unfolded protein response inducers thapsigargin (intracellular calcium releaser) (48) or tunicamycin (glycosylation inhibitor) (47). Exposure to thapsigargin (0.25 μm for 6 h) or tunicamycin (5 μg/ml for 24 h) resulted in a significant increase in phosphorylation of eIF2α and increased protein expression of XBP1 and CHOP (supplemental Fig. S2).
FIGURE 3.
Effect of siRNA-targeted knockdown of HSF-1 on starvation- or heat stress-induced autophagy in A549 cells. A, A549 cells were transfected with either control or HSF-1 siRNA for 48 h. Subsequently, cells were exposed to either control (full media with 100 nm Baf) or starvation media (EBSS with 100 nm Baf) for 2 h. Cells were harvested, lysed, and prepared for Western blot analysis. β-Actin served as an internal control. B represents LC3 autoradiogram quantification based on A. Data represent means ± S.E. (n = 4). *, p < 0.05 versus Scramble siRNA/media; **, p < 0.01 versus Scramble siRNA/media. C, A549 were exposed to heat stress (42 °C for 2 h) or control temperature (37 °C) followed by a 24-h incubation at 37 °C. Next, cells were exposed to either control (full media with 100 nm Baf) or starvation media (EBSS with 100 nm Baf for 2 h). Cells were harvested, lysed, and prepared for Western blot analysis. β-Actin served as an internal control. D represents LC3 autoradiogram quantification based on C. Data represent means ± S.E. (n = 10). *, p < 0.05 versus 37 °C/media; **, p < 0.01 versus 37 ° C/media; ***, p < 0.001 versus AdTrack/media. E, A549 cells were transfected with control or HSF-1 siRNA for 48 h. Next, cells were exposed to control temperature (37 °C) or heat (42 °C) for 2 h with Baf (100 nm). Cells were harvested, lysed, and prepared for Western blot analysis. β-Actin served as an internal control. F represents LC3 autoradiogram quantification based on E. Data represent means ± S.E. (n = 5–6). *, p < 0.05 versus Scramble siRNA/37 °C.
Effect of HSF-1 and HSP70 on Autophagy Regulation
Next, the role of HSF-1 and HSP70 on autophagy was examined. A549 cells were transfected with HSF-1 siRNA to knock down the HSF-1 protein expression followed by infection with adenovirus to overexpress HSP70 protein expression, and their role on starvation- and rapamycin-induced autophagy was tested. As shown in Fig. 4A, HSF-1 siRNA transfection resulted in a marked depletion of HSF-1 expression, and adenovirus infection with Ad70 resulted in a significant increase in HSP70 protein expression in A549 cells. No significant effects of HSP70 overexpression on HSF-1 protein levels were observed. As shown in Fig. 4, A and B, exposure to starvation conditions (EBSS with 100 nm Baf for 2 h) in A549 cells resulted in an increase in autophagy that was inhibited by HSP70 overexpression. Transfection with HSF-1 siRNA potentiated the starvation-induced increase in autophagy, and HSP70 significantly blocked this increase, indicating the strong regulatory role of HSP70 on autophagy. Similarly, transfection with HSF-1 siRNA potentiated the rapamycin-induced increase in autophagy that was prevented by HSP70 overexpression (Fig. 4, C and D).
FIGURE 4.
Effect of siRNA-targeted knockdown of HSF-1 and HSP70 overexpression on starvation- or rapamycin-induced autophagy. A549 cells were transfected with either control or HSF-1 siRNA. After a 6-h incubation, cells were infected with either control (AdTrack) or Ad70 virus (2.5 × 1010 IFUs) to overexpress HSP70. Cells were incubated for the subsequent 3 days. Next, cells were exposed to control conditions (full media with 100 nm Baf) or starvation media (EBSS with 100 nm Baf) for 2 h (A) or rapamycin (50 ng/ml with 100 nm Baf) for 2 h (C). Cells were harvested, lysed, and prepared for Western blot analysis. Densitometric values (B and D) of the corresponding blots (A and C, respectively) were obtained using Photoshop software and normalized to β-actin and set to 1 for control conditions (AdTrack virus, scrambled siRNA, and control culture media). B, represents LC3 autoradiogram quantification based on A. Data represent means ± S.E. (n = 4). *, p < 0.05 versus control conditions (AdTrack virus, scrambled siRNA, and control culture media). **, p < 0.05 versus AdTrack/HSF1 siRNA/control media; #, p < 0.05 versus AdTrack/scrambled siRNA/EBSS; ##, p < 0.05 versus AdTrack/HSF1 siRNA/EBSS. D represents LC3 autoradiogram quantification based on C. Data represent means ± S.E. (n = 7). *, p < 0.05 versus control conditions (AdTrack virus, scrambled siRNA, and control culture media). **, p < 0.01 versus control conditions (AdTrack virus, scrambled siRNA, and control culture media; #, p < 0.05 versus AdTrack virus; HSF1 siRNA, and control culture media.
In the following series of experiments, we further substantiated the regulatory role of HSP70 on macroautophagy in A549 cells by utilizing the recombinant wild type HSP70 and its ATPase domain mutant (K71A). A549 cells were either infected with control (AdTrack), HSP70 overexpressing virus (Ad70), or HSP70 mutant (Ad70M). As shown in Fig. 5, exposure to starvation conditions (EBSS with 100 nm Baf for 2 h) resulted in a significant increase in autophagy in A549 cells. HSP70 overexpression resulted in significant inhibition of this starvation-induced autophagy (Fig. 5, A and F). HSP70 ATPase mutant showed no significant effect on starvation-induced autophagy compared with that observed in starved cells infected with a control HSP70 virus, indicating that HSP70 ATPase domain is important in autophagy regulation.
FIGURE 5.
Effect of HSP70 ATPase mutant on starvation-induced autophagy. A, A549 cells were infected with either control (AdTrack), Ad70, or Ad70 ATPase mutant virus (Ad70M) (2.5 × 1010 IFUs) for 3 days. Subsequently, cells were exposed to either control (full media with 100 nm Baf) or starvation media (EBSS with 100 nm Baf) for 2 h. Cells were harvested, lysed, and prepared for Western blot analysis to measure the relative expression of LC3, HSP70, mTOR, p-mTOR, p-p70S6K, p70S6K Akt, p-Akt, p-4E-BP1, and β-actin (an internal loading control). B–F, data represent means ± S.E. (n = 5–7). B represents p-mTOR to mTOR autoradiogram quantification based on A. *, p < 0.01 versus AdTrack/control media. **, p < 0.05 versus AdTrack/EBSS; ***, p < 0.05 versus Ad70/EBSS. C, represents p-p70S6K to p70S6K autoradiogram quantification based on A. *, p < 0.001 versus AdTrack/control media; **, p < 0.05 versus AdTrack/EBSS; ***, p < 0.05 versus Ad70/EBSS. D represents p-Akt to Akt autoradiogram quantification based on A. #, p < 0.05 versus AdTrack/control media; ##, p < 0.05 versus Ad70/Control media; *, p < 0.05 versus AdTrack/control media; **, p < 0.05 versus AdTrack/EBSS; ***, p < 0.05 versus Ad70/EBSS. E represents p-4E-BP1(γ) to p-4E-BP1(β) autoradiogram quantification based on A. #, p < 0.05 versus AdTrack/control media; ##, p < 0.05 versus Ad70/control media; *, p < 0.05 versus AdTrack/control media; **, p < 0.05 versus AdTrack/EBSS; ***, p < 0.05 versus Ad70/EBSS. F represents LC3-II to β-actin autoradiogram quantification based on A. *, p < 0.001 versus AdTrack/control media; **, p < 0.05 versus AdTrack/EBSS; ***, p < 0.05 versus Ad70/EBSS. G, role of Akt in HSP70-mediated regulation of starvation-induced autophagy. Cells were treated with Akt inhibitor (MK-2206, 3 μm) and HSP70 overexpressing adenovirus (Ad70) (2.5 × 1010 IFUs) for 3 days. Subsequently, cells were exposed to EBSS (with 100 nm Baf) for 2 h. Cells were harvested, lysed, and prepared for Western blot analysis to measure the relative expression of LC3, HSP70, Akt, p-Akt, mTOR, p-mTOR, and β-actin (an internal loading control). H represents p-mTOR autoradiogram quantification based on G. *, p < 0.05 versus AdTrack/DMSO; **, p < 0.01 versus AdTrack/DMSO or Ad70/DMSO. I represents LC3-II autoradiogram quantification based on G. *, p < 0.05 versus AdTrack/DMSO; **, p < 0.01 versus AdTrack/DMSO or Ad70/DMSO, data represent means ± S.E. (n = 3). J, effect of HSP70 ATPase mutant on luciferase refolding in heat-treated A549 cells. Cells transfected with a luciferase expression vector were treated with control (AdTrack), Ad70, or Ad70 ATPase mutant virus (Ad70M) (2.5 × 1010 IFUs) for 3 days. Subsequently cells were heated to 45 °C for 45 min with cycloheximide (10 μg/ml) to prevent de novo HSP70 protein synthesis induced by heat stress. Luciferase levels were measured following a 4-h recovery period at 37 °C. Data represent mean ± S.E. (n = 5). *, p < 0.01 versus AdTrack, **, p < 0.05 versus Ad70.
It has been previously shown that the unfolded protein response triggered by the accumulation of misfolded protein in the ER induces autophagy (49) (50). One potential explanation of our results is that the HSP70-induced inhibition of autophagy was mediated by chaperone activity of HSP70. To test this hypothesis, A549 cells were transfected with pGL.3 to direct a high level of luciferase expression (51) along with control (AdTrack), HSP70 (Ad70), or HSP70 mutant (Ad70M) overexpressing virus. To denature the luciferase, cells were heated (45 °C for 45 min followed by a 4-h recovery period at 37 °C) and harvested, and the luciferase activity was determined. In cells overexpressing HSP70, the luciferase activity was over 2-fold higher than it was in control virus-infected cells, indicating that HSP70 prevented the luciferase activity (Fig. 5J). However, Ad70M did not render that protection, suggesting that HSP70 regulatory role of autophagy is mediated via its chaperone activity.
mTOR signaling plays a crucial role in numerous cellular processes, including cell growth, proliferation, and cell survival (52–55). The kinase mTOR is a main controller of autophagy induction, with inhibited mTOR activating the autophagy (56). We tested mTOR activation by monitoring mTOR phosphorylation (hypophosphorylation of mTOR indicates mTOR inhibition). Exposure of A549 cells to starvation conditions (EBSS with 100 nm Baf for 2 h) resulted in a hypophosphorylation of mTOR (Fig. 5, A and B) that was prevented by overexpression of HSP70 but not its mutant (Fig. 5, A and B). We also studied TOR activity by monitoring phosphorylation of eukaryotic initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) (Fig. 5, A and E), commonly used as an indicator of TOR activation (57, 58). Two isoforms of 4E-BP1 (β and γ) were highly phosphorylated in control cells. Starvation conditions resulted in an increase in the amount of the slowest migrating electrophoretic isoforms (α and β) of 4E-BP1, most probably as a result of hyperphosphorylation. This hyperphosphorylation of the two lowest bands disappeared in cells treated with Ad70. However, in starved cells treated with HSP70 ATPase mutant, the two lowest bands became apparent. Similarly, starvation resulted in a significant decrease in p70S6K phosphorylation (Fig. 5, A and C) that was partially prevented by HSP70 overexpression but not the HSP70 mutant. Considering its important role in autophagy regulation, we hypothesized Akt via activating mTOR (59) to be a target of HSP70 in autophagy regulation. To validate the role of HSP70 on the Akt pathway in autophagy regulation, A549 cells were infected with either control, HSP70 overexpressing virus, or its mutant (Ad70M), and the role of HSP70 on starvation-induced Akt activation was studied. As shown in Fig. 5, A and D, overexpression of HSP70, but not HSP70 ATPase mutant, under both control and starvation conditions, resulted in an increased phosphorylation of Akt, suggesting that HSP70 modulates autophagy through the Akt and mTOR pathway. In the following studies, the role of Akt in HSP70-mediated regulation of starvation-induced autophagy was studied (Fig. 5, G and I). Cells were treated with a highly selective inhibitor of Akt (3 μm MK-2206, Selleck Chemicals, Houston, TX) (60) and HSP70 overexpressing adenovirus (Ad70) (2.5 × 1010 IFUs) for 3 days. Subsequently, cells were exposed to EBSS (with 100 nm Baf) for 2 h. Cells were harvested, lysed, and prepared for Western blot analysis to measure the relative expression of LC3. In cells overexpressing HSP70, we observed an increase in mTOR and Akt phosphorylation that was associated with a decrease in starvation-induced autophagy. Treatment with an Akt inhibitor suppressed mTOR phosphorylation by 30 and 37% under the presence or absence of HSP70 adenovirus exposure, respectively. Exposure to selective Akt inhibitor resulted in a significant increase in autophagy that was not affected by HSP70, indicating that HSP70 exerts its inhibitory effect on autophagy via the Akt pathway.
Under control conditions, autophagy is responsible for the degradation of misfolded and unfolded proteins. Under stress conditions, cells utilize autophagy as a part of homeostatic mechanisms involved in maintaining sufficient macromolecular and ATP production. Although the role of exercise as a stimulus has been recently described in mice (35), the role of exercise on autophagy regulation in humans is less known. In this study, we sought to test the effect of exercise on autophagy regulation in humans under control conditions or glutamine supplementation. Exercise resulted in a significant increase in autophagy in human PBMCs as demonstrated by an increased LC3-II protein expression (Fig. 6, A and B). Gln treatment associated with a significant increase in HSP70 protein expression prevented this exercise-induced increase in autophagy.
FIGURE 6.
Exercise induces autophagy in human PBMCs. In a double blinded study, subjects consumed either placebo or glutamine (0.3 g·kg−1 of fat free mass, three times a day for 7 days). Subjects performed a 60-min treadmill run at 70–80% of the VO2 max in a warm environmental chamber (30 °C). Blood samples were collected before, immediately post, and 2 and 4 h after the exercise. PBMCs were harvested and analyzed for autophagy marker LC3, HSP70, and β-actin (an internal loading control) (A). Data represent means ± S.E. (n = 8). Densitometric values (B and C) of corresponding blot (A) were obtained using Photoshop software and normalized to β-actin and set to 1 for control conditions. B represents LC3-II to β-actin autoradiogram quantification based on A. *, p < 0.05 versus Placebo/Pre; **, p < 0.01 versus Placebo/4 h post exercise. C represents HSP70 to β-actin autoradiogram quantification based on A. *, p < 0.01 versus Placebo/4 h post exercise.
DISCUSSION
Both autophagy and the heat stress response represent protein management alternatives for the stressed cell. Autophagy results in protein and whole organelles degradation (61). Heat shock proteins are generally conservative in nature in that they prevent protein aggregation and facilitate refolding (15, 62). During stress and a subsequent recovery phase, inducible up-regulation of HSP70 protein expression together with other members of the heat shock family allows the cells to survive the noxious conditions and to restore the cellular homeostasis primarily by maintaining proper protein structure (63, 64). Thus, there must be a regulatory system to prioritize one or the other of these protein management systems. In this study, we demonstrate that the heat shock system is prioritized through its ability to interrupt the activation of the autophagic response. In this study, we demonstrate for the first time that overexpression of HSP70 in cell culture models significantly inhibits the starvation-induced autophagy and that this inhibition is associated with an increase in mTOR and Akt activity. Additionally, we show that exercise as a stimulus induced autophagy in humans, and glutamine supplementation associated with an elevated HSP70 protein levels inhibited that exercise-induced autophagy augmentation.
We employed two approaches to test the effect of HSP70 and heat stress response on autophagy regulation. First, we specifically overexpressed HSP70 protein using the adenovirus system whose efficacy was previously proven in both in vitro and in vivo models (30). Overexpression of HSP70 in three cell lines (A549, NCI-H1299, and Caco-2) drastically inhibited starvation-induced autophagy demonstrated by puncta formation or the amount of LC-3 by Western blot analysis. Moreover, under control conditions, HSP70 overexpression resulted in an increased p62 protein expression and p62 puncta formation, indicating decreased autophagy. We then employed an siRNA of HSF-1 to test the effect of heat shock response on autophagy regulation. Knockdown of HSF-1 significantly up-regulated starvation-induced autophagy, suggesting that the absence of one arm of cellular protection leading to accumulation of misfolded proteins results in up-regulation of the other arm of physiological response mainly autophagy. Taken together, our results demonstrate that HSP70 plays a crucial role in autophagy regulation.
Autophagy is a highly controlled process, and mTOR has been shown to play a central in an autophagy modulation. Upstream of mTOR there are several kinases involved in autophagy regulation. Among them, Akt was shown to negatively regulate autophagy. Under growth factor stimulation, an increase in class I PI3K activity blocks and class III PI3K triggers autophagy at the sequestration step (65). Recruitment of Akt to the plasma membrane by PI3K activation is followed by Akt phosphorylation by phosphoinositide-dependent protein kinase (PDK1). Subsequently, Akt phosphorylates and inactivates tuberin, leading to increased mTOR activity (66). Besides playing an important role in autophagy regulation, glucose metabolism, and apoptosis (67), PI3K/Akt was also shown to be important in HSP70 protein expression. Exposure of chronic myelogenous leukemia (K562) cells to resveratrol inhibited Akt activity and resulted in HSF-1 down-regulation and decreased HSP70 protein expression (68). Pharmacological PI3K/Akt inhibitors attenuated HSP70 protein expression, demonstrating the requirement of the PI3K/Akt pathway in HSP70 protein synthesis (69). Similarly, in endothelial cells, Akt overexpression resulted in a marked up-regulation of HSP70. Collectively, those studies demonstrated that the Akt pathway plays a crucial role in HSP70 protein expression. However, the role of HSP70 on the Akt pathway is much less known. In recent studies, it has been shown that HSP70 ATPase activators increased and inhibitors decreased Akt levels in HeLa cells (70). In this study, we demonstrated that HSP70 overexpression inhibited starvation-induced autophagy, and this inhibition was associated with an up-regulation of the mTOR/Akt pathway, suggesting that HSP70-induced Akt phosphorylation led to mTOR phosphorylation resulting in a blocked autophagy. Future studies are needed to fully validate the role of HSP70 in PI3K/Akt/mTOR pathway.
Endoplasmic reticulum is an intracellular organelle responsible for protein folding into their native structures as well as their post-translational modifications, including glycosylation and disulfide bond formation (71). Under various physiological (increased secretory load) or pathological conditions (accumulation of mutated proteins) folding or protein assembly is compromised leading to up-regulation of unfolded protein response (72). The ER quality control mechanism, including up-regulation of chaperone proteins, ensures that only properly folded proteins can be transported out of the ER, otherwise they get degraded in the cytosol by a process called ER-associated protein degradation (73). If the stress sustains, the compensatory mechanisms get overwhelmed leading to apoptosis or cell death (48, 74). It has been shown that heat stress, oxidative stress, and heavy metal induce ER stress response (75). Exposure of colon and prostate cancer cells to ER stress inducers resulted in a significant up-regulation of autophagy. In this study, heat stress associated with an increase in HSP70 resulted in an autophagy augmentation (Fig. 3, C and D). Although the direct answer cannot be offered, we suggest that heat stress induces unfolded protein response (49), not seen under starvation or rapamycin treatment, subsequently leading to exhaustion of HSP70 folding capacity and finally increased autophagy representing an adaptive response responsible for unfolded/misfolded protein clearance (74). Previous studies have suggested that ER stress is involved in the development of many pathological conditions, including Huntington and Parkinson diseases, and insulin resistance and inhibition of ER stress seemed to be therapeutically beneficial in numerous cell culture and animal models (76–81). Recently, it has been shown that ER stress results in autophagy induction (50), and induction of autophagy plays a cytoprotective role during the unfolded protein response (82). However, the role of autophagy in cell death or survival remains a matter of debate. It appears that the role of autophagy in cell fate, death or survival, is highly cell type- and stress nature-dependent (83), and this autophagy-associated cell death was prevented by silencing the autophagy genes (84). Our results show that HSP70 overexpression represents a new component in an already very complex network of autophagy regulation. Similarly, inflammation represents a system of detection and reaction to dangerous signals such as pathogens, damaged cells, injured tissue, or irritants (85). Inflammatory response is highly controlled and regulated. The right level of inflammatory response is fundamental to the survival of the organism. However, prolonged and exaggerated inflammatory response may be detrimental or even deadly. In numerous cell culture and animal models, heat shock proteins have been shown to modulate inflammatory response. Heat pre-conditioning, severe enough to up-regulate HSP70 protein expression, protected animals against the lethal effect of endotoxin (24, 25). Glutamine administration, associated with HSP70 protein expression, improved survival in rat sepsis (86). We have shown recently that the effect of heat pre-conditioning on modulating the inflammatory response can be mimicked by HSP70 overexpression. In rats, LPS-induced cytokine expression was inhibited by HSP70 overexpression, and this inhibition was associated with a blockade of the NF-κB pathway (30) by preventing NF-κB p65 nuclear translocation and IκBα degradation. We speculate that heat shock response, in terms of regulation of both autophagy and inflammation, plays a regulatory role in fine adjustments when they get out of control.
Previously, it has been shown that exercise resulted in an increase in autophagy in mice (35). Its role was shown to be required for muscle glucose homeostasis. Using our human exercise protocol, we have now demonstrated that exercise associated with an increase (≥39 °C) in body temperature resulted in a significant increase in autophagy measured in PBMCs. These in vivo data support our in vitro findings showing that autophagy is increased under hyperthermic conditions. Glutamine supplementation as a conditioning stimulus prior to exercise led to increased levels of HSP70 and prevented exercise-induced autophagy in PBMCs, indicating that HSP70 regulates stress-induced autophagy in humans. The exact role of glutamine in regulation of autophagy is complex and multifactorial. Glutamine deprivation in perfused rat liver resulted in a significant increase in autophagy (87–89). In contrast, in intestinal epithelial cells, glutamine administration increased autophagy under basal and stress conditions (75). Recently, it has been shown that FOXO-induced glutamine production in the Ba/F3 (a murine interleukin-3 dependent pro-B) cell line resulted in mTOR inhibition and an increased level of autophagy (90). Finally, in co-culture, glutamine exposure led to an increase in autophagy markers in fibroblasts and inhibited autophagy in MCF7 breast cancer cells (91). Several cellular mechanisms of glutamine on autophagy regulation have been indicated, including p38 MAPK (75), TIGER (91), and FOXO (90). This study was designed to use glutamine supplementation to increase cellular HSP70 prior to the exercise stress to provide a human counterpart to our cell-based studies. As predicted by these cell studies, the accumulation of HSP70 prior to exercise inhibited stress-induced autophagy in exercising humans. Further studies are needed to fully determine cellular and molecular mechanisms of glutamine and other heat shock response modulators on exercise-induced autophagy.
In summary, we report for the first time that HSP70 overexpression inhibits and HSF1 knockdown increases autophagy in cell culture model. Second, HSP70 may modulate autophagy by regulating mTOR/Akt pathway. Our studies also show that exercise associated with hyperthermia induced autophagy in humans that is prevented by glutamine-induced HSP70. Collectively, our data indicate that autophagy involved in maintaining energy and protein homeostasis remains under the regulatory control of HSP70.
Supplementary Material
Acknowledgments
We thank Dr. W. Wharton and R. Lobb for their excellent technical and professional assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants RM-07-007.

This article contains supplemental Figs. S1 and S2.
- HSP
- heat shock protein
- EBSS
- Earle's balanced salt solution
- mTOR
- mammalian target of rapamycin
- ER
- endoplasmic reticulum
- PBMC
- peripheral blood mononuclear cell
- Baf
- bafilomycin A1
- IFU
- infectious unit.
REFERENCES
- 1. Mizushima N., Levine B. (2010) Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kroemer G., Mariño G., Levine B. (2010) Autophagy and the integrated stress response. Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizushima N., Yoshimori T., Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 4. Rubinsztein D. C., Codogno P., Levine B. (2012) Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levine B., Deretic V. (2007) Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 7, 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ravikumar B., Futter M., Jahreiss L., Korolchuk V. I., Lichtenberg M., Luo S., Massey D. C., Menzies F. M., Narayanan U., Renna M., Jimenez-Sanchez M., Sarkar S., Underwood B., Winslow A., Rubinsztein D. C. (2009) Mammalian macroautophagy at a glance. J. Cell Sci. 122, 1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren Y., Huang F., Liu Y., Yang Y., Jiang Q., Xu C. (2009) Autophagy inhibition through PI3K/Akt increases apoptosis by sodium selenite in NB4 cells. BMB Rep. 42, 599–604 [DOI] [PubMed] [Google Scholar]
- 8. Criollo A., Senovilla L., Authier H., Maiuri M. C., Morselli E., Vitale I., Kepp O., Tasdemir E., Galluzzi L., Shen S., Tailler M., Delahaye N., Tesniere A., De Stefano D., Younes A. B., Harper F., Pierron G., Lavandero S., Zitvogel L., Israel A., Baud V., Kroemer G. (2010) The IKK complex contributes to the induction of autophagy. EMBO J. 29, 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ogier-Denis E., Pattingre S., El Benna J., Codogno P. (2000) Erk1/2-dependent phosphorylation of Gα-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J. Biol. Chem. 275, 39090–39095 [DOI] [PubMed] [Google Scholar]
- 10. Pattingre S., Bauvy C., Codogno P. (2003) Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J. Biol. Chem. 278, 16667–16674 [DOI] [PubMed] [Google Scholar]
- 11. Aoki H., Takada Y., Kondo S., Sawaya R., Aggarwal B. B., Kondo Y. (2007) Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol. Pharmacol. 72, 29–39 [DOI] [PubMed] [Google Scholar]
- 12. Meley D., Bauvy C., Houben-Weerts J. H., Dubbelhuis P. F., Helmond M. T., Codogno P., Meijer A. J. (2006) AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 281, 34870–34879 [DOI] [PubMed] [Google Scholar]
- 13. Feng Z., Zhang H., Levine A. J., Jin S. (2005) The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. U.S.A. 102, 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tasdemir E., Maiuri M. C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'Amelio M., Criollo A., Morselli E., Zhu C., Harper F., Nannmark U., Samara C., Pinton P., Vicencio J. M., Carnuccio R., Moll U. M., Madeo F., Paterlini-Brechot P., Rizzuto R., Szabadkai G., Pierron G., Blomgren K., Tavernarakis N., Codogno P., Cecconi F., Kroemer G. (2008) Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 10, 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moseley P. L., Gapen C., Wallen E. S., Walter M. E., Peterson M. W. (1994) Thermal stress induces epithelial permeability. Am. J. Physiol. 267, C425–C434 [DOI] [PubMed] [Google Scholar]
- 16. Gething M. J., Sambrook J. (1992) Protein folding in the cell. Nature 355, 33–45 [DOI] [PubMed] [Google Scholar]
- 17. Hendrick J. P., Hartl F. U. (1993) Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 62, 349–384 [DOI] [PubMed] [Google Scholar]
- 18. Parsell D. A., Lindquist S. (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496 [DOI] [PubMed] [Google Scholar]
- 19. Hartl F. U. (1996) Molecular chaperones in cellular protein folding. Nature 381, 571–579 [DOI] [PubMed] [Google Scholar]
- 20. Mosser D. D., Martin L. H. (1992) Induced thermotolerance to apoptosis in a human T lymphocyte cell line. J. Cell Physiol. 151, 561–570 [DOI] [PubMed] [Google Scholar]
- 21. Mailhos C., Howard M. K., Latchman D. S. (1993) Heat shock protects neuronal cells from programmed cell death by apoptosis. Neuroscience 55, 621–627 [DOI] [PubMed] [Google Scholar]
- 22. Musch M. W., Ciancio M. J., Sarge K., Chang E. B. (1996) Induction of heat shock protein 70 protects intestinal epithelial IEC-18 cells from oxidant and thermal injury. Am. J. Physiol. 270, C429–C436 [DOI] [PubMed] [Google Scholar]
- 23. Murata M., Gong P., Suzuki K., Koizumi S. (1999) Differential metal response and regulation of human heavy metal-inducible genes. J. Cell Physiol. 180, 105–113 [DOI] [PubMed] [Google Scholar]
- 24. Ryan A. J., Flanagan S. W., Moseley P. L., Gisolfi C. V. (1992) Acute heat stress protects rats against endotoxin shock. J. Appl. Physiol. 73, 1517–1522 [DOI] [PubMed] [Google Scholar]
- 25. Hotchkiss R., Nunnally I., Lindquist S., Taulien J., Perdrizet G., Karl I. (1993) Hyperthermia protects mice against the lethal effects of endotoxin. Am. J. Physiol. 265, R1447–R1457 [DOI] [PubMed] [Google Scholar]
- 26. Chu E. K., Ribeiro S. P., Slutsky A. S. (1997) Heat stress increases survival rates in lipopolysaccharide-stimulated rats. Crit. Care Med. 25, 1727–1732 [DOI] [PubMed] [Google Scholar]
- 27. Dokladny K., Wharton W., Lobb R., Ma T. Y., Moseley P. L. (2006) Induction of physiological thermotolerance in MDCK monolayers: contribution of heat shock protein 70. Cell Stress Chaperones 11, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dokladny K., Moseley P. L., Ma T. Y. (2006) Physiologically relevant increase in temperature causes an increase in intestinal epithelial tight junction permeability. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G204–G212 [DOI] [PubMed] [Google Scholar]
- 29. Dokladny K., Ye D., Kennedy J. C., Moseley P. L., Ma T. Y. (2008) Cellular and molecular mechanisms of heat stress-induced up-regulation of occludin protein expression: regulatory role of heat shock factor-1. Am. J. Pathol. 172, 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dokladny K., Lobb R., Wharton W., Ma T. Y., Moseley P. L. (2010) LPS-induced cytokine levels are repressed by elevated expression of HSP70 in rats: possible role of NF-κB. Cell Stress Chaperones 15, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kluger M. J., Rudolph K., Soszynski D., Conn C. A., Leon L. R., Kozak W., Wallen E. S., Moseley P. L. (1997) Effect of heat stress on LPS-induced fever and tumor necrosis factor. Am. J. Physiol. 273, R858–R863 [DOI] [PubMed] [Google Scholar]
- 32. Dokladny K., Kozak A., Wachulec M., Wallen E. S., Menache M. G., Kozak W., Kluger M. J., Moseley P. L. (2001) Effect of heat stress on LPS-induced febrile response in d-galactosamine-sensitized rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280, R338–R344 [DOI] [PubMed] [Google Scholar]
- 33. Qin L., Wang Z., Tao L., Wang Y. (2010) ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6, 239–247 [DOI] [PubMed] [Google Scholar]
- 34. Park M. A., Yacoub A., Rahmani M., Zhang G., Hart L., Hagan M. P., Calderwood S. K., Sherman M. Y., Koumenis C., Spiegel S., Chen C. S., Graf M., Curiel D. T., Fisher P. B., Grant S., Dent P. (2008) OSU-03012 stimulates PKR-like endoplasmic reticulum-dependent increases in 70-kDa heat shock protein expression, attenuating its lethal actions in transformed cells. Mol. Pharmacol. 73, 1168–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He C., Bassik M. C., Moresi V., Sun K., Wei Y., Zou Z., An Z., Loh J., Fisher J., Sun Q., Korsmeyer S., Packer M., May H. I., Hill J. A., Virgin H. W., Gilpin C., Xiao G., Bassel-Duby R., Scherer P. E., Levine B. (2012) Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He C., Sumpter R., Jr., Levine B. (2012) Exercise induces autophagy in peripheral tissues and in the brain. Autophagy 8, 1548–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohsumi Y., Mizushima N. (2004) Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 15, 231–236 [DOI] [PubMed] [Google Scholar]
- 39. Vergne I., Roberts E., Elmaoued R. A., Tosch V., Delgado M. A., Proikas-Cezanne T., Laporte J., Deretic V. (2009) Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 28, 2244–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Droge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fesus L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., Gonzalez-Estevez C., Gorski S., Gottlieb R. A., Haussinger D., He Y. W., Heidenreich K., Hill J. A., Hoyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jaattela M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovacs A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., Lopez-Otin C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Melendez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Munz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nurnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Talloczy Z., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcategui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E. M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 45. Morimoto R. I. (1993) Cells in stress: transcriptional activation of heat shock genes. Science 259, 1409–1410 [DOI] [PubMed] [Google Scholar]
- 46. Sarge K. D., Murphy S. P., Morimoto R. I. (1993) Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13, 1392–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bolt A. M., Zhao F., Pacheco S., Klimecki W. T. (2012) Arsenite-induced autophagy is associated with proteotoxicity in human lymphoblastoid cells. Toxicol. Appl. Pharmacol. 264, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gupta S., Deepti A., Deegan S., Lisbona F., Hetz C., Samali A. (2010) HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1α-XBP1 signaling through a physical interaction. PLoS Biol. 8, e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Patil C., Walter P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 [DOI] [PubMed] [Google Scholar]
- 50. Yorimitsu T., Nair U., Yang Z., Klionsky D. J. (2006) Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dokladny K., Wharton W., Ma T. Y., Moseley P. L. (2008) Lack of cross-tolerance following heat and cadmium exposure in functional MDCK monolayers. J. Appl. Toxicol. 28, 885–894 [DOI] [PubMed] [Google Scholar]
- 52. Yu J., Yaba A., Kasiman C., Thomson T., Johnson J. (2011) mTOR controls ovarian follicle growth by regulating granulosa cell proliferation. PLoS One 6, e21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salmond R. J., Zamoyska R. (2011) The influence of mTOR on T helper cell differentiation and dendritic cell function. Eur. J. Immunol. 41, 2137–2141 [DOI] [PubMed] [Google Scholar]
- 54. Machado-Neto J. A., Favaro P., Lazarini M., Costa F. F., Olalla Saad S. T., Traina F. (2011) Knockdown of insulin receptor substrate 1 reduces proliferation and down-regulates Akt/mTOR and MAPK pathways in K562 cells. Biochim. Biophys. Acta 1813, 1404–1411 [DOI] [PubMed] [Google Scholar]
- 55. Li Q., Rao R. R., Araki K., Pollizzi K., Odunsi K., Powell J. D., Shrikant P. A. (2011) A central role for mTOR kinase in homeostatic proliferation-induced CD8+ T cell memory and tumor immunity. Immunity 34, 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liang C. (2010) Negative regulation of autophagy. Cell Death Differ. 17, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 58. Beretta L., Gingras A. C., Svitkin Y. V., Hall M. N., Sonenberg N. (1996) Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 15, 658–664 [PMC free article] [PubMed] [Google Scholar]
- 59. Potter C. J., Pedraza L. G., Xu T. (2002) Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4, 658–665 [DOI] [PubMed] [Google Scholar]
- 60. Hirai H., Sootome H., Nakatsuru Y., Miyama K., Taguchi S., Tsujioka K., Ueno Y., Hatch H., Majumder P. K., Pan B. S., Kotani H. (2010) MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer Ther. 9, 1956–1967 [DOI] [PubMed] [Google Scholar]
- 61. Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Self-eating and self-killing: cross-talk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 62. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Craig E. A., Weissman J. S., Horwich A. L. (1994) Heat shock proteins and molecular chaperones: mediators of protein conformation and turnover in the cell. Cell 78, 365–372 [DOI] [PubMed] [Google Scholar]
- 64. Liang P., MacRae T. H. (1997) Molecular chaperones and the cytoskeleton. J. Cell Sci. 110, 1431–1440 [DOI] [PubMed] [Google Scholar]
- 65. Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000) Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992–998 [DOI] [PubMed] [Google Scholar]
- 66. Shaw R. J., Cantley L. C. (2006) Ras, PI(3)K, and mTOR signalling controls tumour cell growth. Nature 441, 424–430 [DOI] [PubMed] [Google Scholar]
- 67. Franke T. F. (2008) PI3K/Akt: getting it right matters. Oncogene 27, 6473–6488 [DOI] [PubMed] [Google Scholar]
- 68. Banerjee Mustafi S., Chakraborty P. K., Raha S. (2011) Modulation of Akt and ERK1/2 pathways by resveratrol in chronic myelogenous leukemia (CML) cells results in the down-regulation of Hsp70. PLoS One 5, e8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou J., Schmid T., Frank R., Brüne B. (2004) PI3K/Akt is required for heat shock proteins to protect hypoxia-inducible factor 1α from pVHL-independent degradation. J. Biol. Chem. 279, 13506–13513 [DOI] [PubMed] [Google Scholar]
- 70. Koren J., 3rd, Jinwal U. K., Jin Y., O'Leary J., Jones J. R., Johnson A. G., Blair L. J., Abisambra J. F., Chang L., Miyata Y., Cheng A. M., Guo J., Cheng J. Q., Gestwicki J. E., Dickey C. A. (2010) Facilitating Akt clearance via manipulation of Hsp70 activity and levels. J. Biol. Chem. 285, 2498–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lin J. H., Walter P., Yen T. S. (2008) Endoplasmic reticulum stress in disease pathogenesis. Annu. Rev. Pathol. 3, 399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scriven P., Brown N. J., Pockley A. G., Wyld L. (2007) The unfolded protein response and cancer: a brighter future unfolding? J. Mol. Med. 85, 331–341 [DOI] [PubMed] [Google Scholar]
- 73. Merksamer P. I., Papa F. R. (2010) The UPR and cell fate at a glance. J. Cell Sci. 123, 1003–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ding W. X., Ni H. M., Gao W., Hou Y. F., Melan M. A., Chen X., Stolz D. B., Shao Z. M., Yin X. M. (2007) Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 282, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 75. Sakiyama T., Musch M. W., Ropeleski M. J., Tsubouchi H., Chang E. B. (2009) Glutamine increases autophagy under basal and stressed conditions in intestinal epithelial cells. Gastroenterology 136, 924–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Reijonen S., Putkonen N., Nørremølle A., Lindholm D., Korhonen L. (2008) Inhibition of endoplasmic reticulum stress counteracts neuronal cell death and protein aggregation caused by N-terminal mutant huntingtin proteins. Exp. Cell Res. 314, 950–960 [DOI] [PubMed] [Google Scholar]
- 77. Atwal R. S., Truant R. (2008) A stress sensitive ER membrane-association domain in Huntingtin protein defines a potential role for Huntingtin in the regulation of autophagy. Autophagy 4, 91–93 [DOI] [PubMed] [Google Scholar]
- 78. Pereira C., Ferreiro E., Cardoso S. M., de Oliveira C. R. (2004) Cell degeneration induced by amyloid-β peptides: implications for Alzheimer's disease. J. Mol. Neurosci. 23, 97–104 [DOI] [PubMed] [Google Scholar]
- 79. Zhang Y., Dong L., Yang X., Shi H., Zhang L. (2011) α-Linolenic acid prevents endoplasmic reticulum stress-mediated apoptosis of stearic acid lipotoxicity on primary rat hepatocytes. Lipids Health Dis. 10, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gregor M. F., Hotamisligil G. S. (2007) Thematic review series: Adipocyte Biology. Adipocyte stress: the endoplasmic reticulum and metabolic disease. J. Lipid Res. 48, 1905–1914 [DOI] [PubMed] [Google Scholar]
- 81. Yang L., Li P., Fu S., Calay E. S., Hotamisligil G. S. (2010) Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bernales S., McDonald K. L., Walter P. (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Baehrecke E. H. (2005) Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6, 505–510 [DOI] [PubMed] [Google Scholar]
- 84. Kroemer G., Levine B. (2008) Autophagic cell death: the story of a misnomer. Nat. Rev. Mol. Cell Biol. 9, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Moseley P. L. (1998) Heat shock proteins and the inflammatory response. Ann. N.Y. Acad. Sci. 856, 206–213 [DOI] [PubMed] [Google Scholar]
- 86. Kwon W. Y., Suh G. J., Kim K. S., Jo Y. H., Lee J. H., Kim K., Jung S. K. (2010) Glutamine attenuates acute lung injury by inhibition of high mobility group box protein-1 expression during sepsis. Br. J. Nutr. 103, 890–898 [DOI] [PubMed] [Google Scholar]
- 87. Jardon M. A., Sattha B., Braasch K., Leung A. O., Côté H. C., Butler M., Gorski S. M., Piret J. M. (2012) Inhibition of glutamine-dependent autophagy increases t-PA production in CHO cell fed-batch processes. Biotechnol. Bioeng. 109, 1228–1238 [DOI] [PubMed] [Google Scholar]
- 88. Mortimore G. E., Schworer C. M. (1977) Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 270, 174–176 [DOI] [PubMed] [Google Scholar]
- 89. Pösö A. R., Wert J. J., Jr., Mortimore G. E. (1982) Multifunctional control of amino acids of deprivation-induced proteolysis in liver. Role of leucine. J. Biol. Chem. 257, 12114–12120 [PubMed] [Google Scholar]
- 90. van der Vos K. E., Eliasson P., Proikas-Cezanne T., Vervoort S. J., van Boxtel R., Putker M., van Zutphen I. J., Mauthe M., Zellmer S., Pals C., Verhagen L. P., Groot Koerkamp M. J., Braat A. K., Dansen T. B., Holstege F. C., Gebhardt R., Burgering B. M., Coffer P. J. (2012) Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat. Cell Biol. 14, 829–837 [DOI] [PubMed] [Google Scholar]
- 91. Ko Y. H., Lin Z., Flomenberg N., Pestell R. G., Howell A., Sotgia F., Lisanti M. P., Martinez-Outschoorn U. E. (2011) Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance. Cancer Biol. Ther. 12, 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.