Abstract
Polyubiquitin (polyUb) chain topology is thought to direct modified substrates to specific fates, but this function-topology relationship is poorly understood, as are the dynamics and subcellular locations of specific polyUb signals. Experimental access to these questions has been limited because linkage-specific inhibitors and in vivo sensors have been unavailable. Here we present a general strategy to track linkage-specific polyUb signals in yeast and mammalian cells, and to probe their functions. We designed several high-affinity lysine-63-polyUb-binding proteins and demonstrate their specificity both in vitro and in cells. We apply these tools as competitive inhibitors to dissect the polyUb-linkage dependence of NF-κB activation in several cell types, inferring the essential role of lysine-63-polyUb for signaling via the IL-1β and TNF-related weak inducer of apoptosis (TWEAK) but not TNF-α receptors. We anticipate live-cell imaging, proteomic, and biochemical applications for these tools, and extension of the design strategy to other polymeric ubiquitin-like protein modifications.
INTRODUCTION
The small protein-modifier ubiquitin (Ub) is an essential signaling element in degradative and non-degradative processes that span nearly all of eukaryotic cell biology1. This functional diversity is explained in part by the structural diversity of Ub modifications. Many forms of polyubiquitin (polyUb) are present in cells, each distinguished by the Ub–Ub linkages along the chain1. Ub has seven lysines and an α-amine that can each accept a Ub carboxy-terminus in an (iso)peptide bond for chain formation2. The prevailing model holds that each structurally unique linkage-type can recruit a distinct set of effectors that influence the fate of the modified protein1, 3,4.
Mapping polyUb topologies to functions inside cells is critical to validate this model and fully understand Ub-dependent regulation. However, assessing this relationship has been particularly difficult. High endogenous expression levels and a multi-locus gene structure make manipulation of Ub pools in vivo cumbersome or impossible outside of a few model systems like yeast5,6 or specially-constructed cell lines7. Linkage-specific probes for live cells have not been available. In vitro, different polyUb linkages can be identified by mass spectrometric methods2, but like many post-translational modifications, polyubiquitination is highly dynamic, and low abundance of polyUb conjugates has limited application of these approaches. Thus, though much of the proteome is subject to polyubiquitination, little is known about how linkage-type relates to the functions and fates of those modified proteins1,4. Evidence supports that polyUb assembled through lysine-48 (Lys48-polyUb) is a dedicated signal for proteasome recruitment and substrate degradation throughout the cell8, whereas Lys63-polyUb serves non-proteasomal roles in DNA repair9,10, autophagy and organelle clearance11, endocytosis12, and inflammation signaling13. Even for these examples, the degree to which other polyUb topologies may also serve these functions, the dynamics of these signals in cells, and the relationship between subcellular localization and signaling fidelity are essentially unknown and difficult to examine with current methods.
To address this deficit, we set out to develop sensors that could be expressed in cells to localize or inhibit specific polyUb signals. Building on structural analyses and principles of linkage-specific avidity14,15, we created high-affinity polyUb-binding proteins from multiple tandem Ubiquitin Interacting Motifs (tUIMs) with structured linker regions that direct binding to Lys63-polyUb. These proteins localized to Lys63-specific structures when expressed in cells, and can be applied as inhibitors to diagnose the linkage-type dependence of polyUb signaling in virtually any cell type.
RESULTS
Avidity-based design for linkage-specific polyUb binding
We previously described how the short linkers between domains in naturally-occurring tUIM proteins can determine polyUb linkage specificity by promoting avid interactions with multiple Ub units in a particular configuration14. Helical 7-amino acid linkers position 2-UIM peptides for optimum avid binding across a single Lys63-linkage, but not to other linkage-types or monoUb. Whereas 8-amino acid linkers are near-optimum for Lys63 binding, 6-amino acid helical linkers confer low affinities for Lys63-polyUb because they position the Ub-binding sites out of helical phase with each other (Fig. 1a,b). We extended this natural strategy to create “Vx3”, a 3-UIM peptide with the relatively tight-binding Vps27 UIM domains (KdmonoUb = 100–200 μM16) and weakly helical 7-amino acid linkers. Remarkably, Vx3 bound Lys63-polyUb with Kd = 4.4 nM, a 70-fold preference over Lys48-polyUb, and a 2,700-fold preference over linear (α-amine-linked) polyUb.
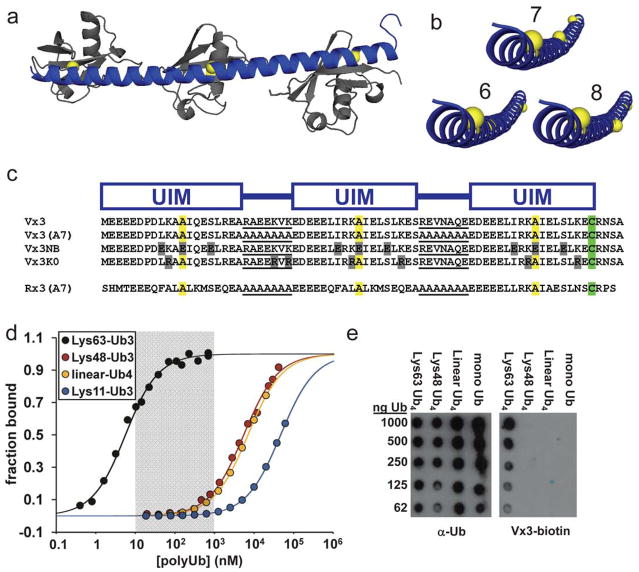
Figure 1. An avidity-based design strategy yields tUIM peptides with high affinity and linkage specificity.
(a) A homology model of a 3-UIM peptide (blue) bound to Lys63-Ub3 (gray) was created by aligning a continuous α-helical segment to the UIM–monoUb complex structure for Vsp2720 and positioning additional Ub units along the Lys63–C-terminus axis. α-Helical, 7-amino acid linkers should orient UIM binding sites (yellow spheres) in phase for optimal contact across all three Ub units. (b) Comparison of the 3-UIM peptide depicted in (a) with versions containing 6- or 8-amino acid α-helical linkers. 8-Amino acid linkers should be near optimum for Lys63-specific avidity, but 6-residue linkers put binding sites far out of helical phase. (c) 3-UIM peptides designed for this study are shown highlighting the arrangement of UIMs, linker sequences (underlined), conserved alanines (yellow) at the centers of Ub binding sites, and the cysteine residues used for labeling (green). Key residue differences are in gray. (d) Fluorescence anisotropy binding data for Rx3(A7)-fluorescein titrated with purified polyUb chains linked through Ub Lys63 (black), Lys48 (red), the N-terminal αamine (“linear”, yellow), or Lys11 (blue). (e) Vx3-biotin or a non-linkage-specific Ub antibody were used to probe membranes spotted with purified (poly)Ub species. See also Supplementary Fig. 1.
Consistent with the principle that more-structured linkers provide higher affinity and specificity, strongly α-helical hepta-alanine linkers (Supplementary Fig. 1) improved the affinity and specificity of Vx3 (Table 1, Vx3(A7)). This peptide bound to Lys63-Ub4 more tightly than we could accurately measure, with Kd ≤ 200 pM (Supplementary Fig. 1). In spite of the improved Lys63-linkage specificity, we reasoned that the range of concentrations over which Vx3(A7) was highly linkage-specific (0.01 – 1 nM) was too low to be useful in cells because physiological concentrations of non- Lys63-linked species probably exceed 1 nM in some cases; total cellular Ub may range several-fold to 85 μM, with ~10% distributed among mostly Lys48-, Lys63-, and Lys11-linked polyUb conjugates2,17.
Table 1.
Equilibrium dissociation constants (Kd) and specificities for designed tUIM peptides interacting with purified tri-or tetraUb chains.a
| tUIM peptide | K63-polyUb Ub3 (Ub4) (nM) | K48-polyUb Ub3 (Ub4) (nM) | K11-Ub3 (nM) | linear-Ub4 (nM) | K63/K48 preference | K63/K11 preference | K63/linear preference |
|---|---|---|---|---|---|---|---|
| Vx3 | 7.4 (4.4) | 540 (310) | 9,000 | ~70-fold | >2,700-fold | ||
| Vx3K0 | 5.7 (4.1) | 730 | ~130-fold | ||||
| Vx3(A7) | (<0.2) | (32.0) | 4,000 | >160-fold | >20,000- old | ||
| Rx3(A7) | 5.0 (3.0) | 5,400 | >10,000b | 6,900 | ~1,100-fold | >3,300-fold | >23,000- old |
All values were measured by fluorescence anisotropy. Standard deviations typically were < 5%, and replicates typically differed by <10%.
The best-fit value of a non-saturating titration was 46,000 nM; 10,000 nM is an estimate of the lower limit of this equilibrium constant.
To shift the high-specificity regime to high-nanomolar concentrations, we paired the weaker UIMs from human Rap80 (KdmonoUb = 500–1,000 μM14) with all-alanine linkers to create Rx3(A7) (Fig. 1c). Indeed, Rx3(A7) bound Lys63-polyUb as tightly as Vx3 (KdUb3 = 5 nM) but with additional specificity conferred by the highly-structured linkers. We measured a 1,100-fold preference for Lys63- over Lys48-polyUb, and a 23,000-fold preference over linear-polyUb. Binding to Lys11-polyUb was too weak for us to saturate, but the preference for Lys63- over Lys11-polyUb probably exceeds 3,300-fold (Table 1; Fig. 1d). Rx3(A7) therefore displays high linkage-specificity for Lys63-polyUb over a broad concentration range that likely encompasses typical cellular polyUb levels (shaded region, Fig. 1d). Although this range is narrower for Vx3 (see Table 1), as described below, in many applications Rx3 and Vx3 can be used with qualitatively similar effects. As expected from structural modeling (Fig. 1b), an 8-alanine linker conferred weak Lys63-polyUb binding in the 3-UIM context (Rx3(A8)), and Rx3(A6) had even lower affinity (Kd = 160 nM and 2,900 nM, respectively; Supplementary Fig. 1).
These results indicated that linkage-specific avidity may be applied to produce polyUb-binding reagents of arbitrarily high affinity and with linkage specificities that rival or exceed linkage-specific polyUb antibodies18,19. Indeed, biotin-labeled Vx3 bound specifically to Lys63-polyUb chains immobilized on a membrane, and thus may be used to identify polyUb types much like an antibody in immunoblot and other biochemical applications (Fig. 1e). Unlike antibodies, however, these reagents may be easily expressed inside living cells; here we focus on these applications.
Intracellular Lys63-linked polyUb structures and dynamics
We transiently expressed tUIM fusions to mEGFP (monomeric Enhanced Green Fluorescent Protein) in cultured human cells. Both Vx3-mEGFP and Rx3(A7)-mEGFP were somewhat excluded from the nucleus and localized to large perinuclear foci that vary widely in appearance from cell to cell (Fig. 2a; Supplementary Fig. 2). Similar structures have been observed in cells examined by immunofluorescence with anti-Lys63-polyUb antibodies19. A subset of these foci co-localized with markers of autophagic vesicles (Supplementary Fig. 2), consistent with recent reports of a role for Lys63-polyUb in cargo selection for some autophagy processes11. Consistent with Lys63-polyUb binding, the intermediate Lys63-binder Rx3(A8)-mEGFP was mostly diffuse, with cytosolic punctae appearing only in cells that expressed the very highest levels of the fusion protein. The weaker-binding Rx3(A6)-mEGFP was completely diffuse in all cells, identical to the non-Ub-binding control protein Vx3NB-mEGFP (Figs. 1c and 2b,d–f). Nuclear-localized NLS-Vx3-EGFP appeared in smaller and more uniformly-sized foci (Fig. 2c), a subset of which co-localized with the DNA double strand break (DSB) markerγH2AX (Fig. 4d, and discussed below). We found that some Vx3-EGFP was apparently ubiquitinated in human cells (≤10%, data not shown), an unwanted modification that could be eliminated by lysine-to-arginine substitutions within the tUIM sequence. The thus altered peptide, Vx3K0, was virtually indistinguishable from Vx3 with respect to in vitro polyUb affinity (Table 1), subcellular localization, and inhibitory activity (discussed below). Taken together, these results indicate that the cellular structures revealed by the Vx3 and Rx3(A7) proteins were ubiquitin-specific, but were not the result of a generic Ub-binding capacity. Instead, these peptides appeared to localize to cellular structures through interactions with specific polyUb topologies.
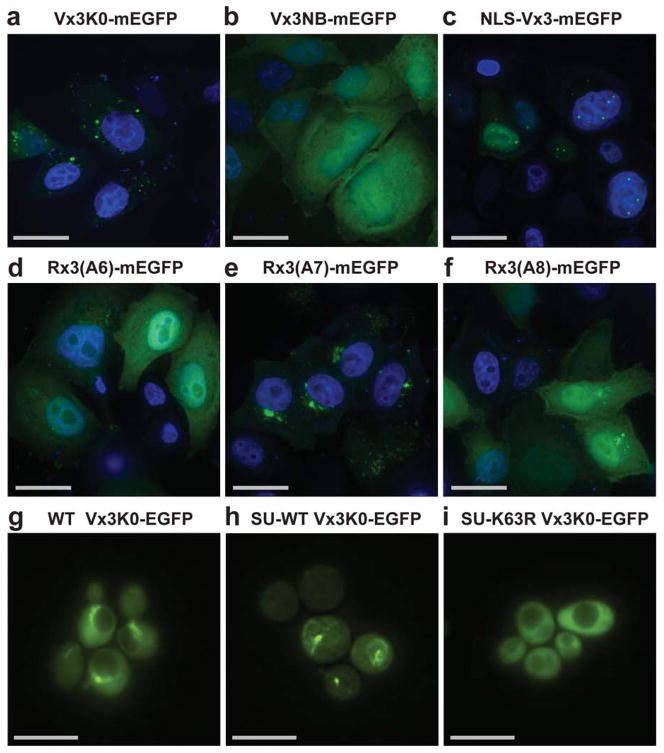
Figure 2. Designed proteins localize to linkage-specific structures inside cells.
Images are shown for HeLa cells expressing mEGFP fusions to indicated tUIM peptides with DAPI DNA counterstaining (a–f). Wild-type (WT) (g), or Single-Ub yeast strains (SU) expressing wild-type Ub (SU-WT) (h) or Lys63Arg-mutated Ub (SU-K63R) (i) together with Vx3K0-EGFP are shown. Scale bars, 30 μm (a–f) and 10 μm (g–i). See also Supplementary Fig. 2.
Figure 4. tUIM sensor proteins can inhibit linkage-specific functions of cellular polyUb.
(a) HeLa cell lysates prepared from transiently transfected cells expressing Rx3(A7)-mEGFP or the non-Ub-binding control Vx3NB-mEGFP were probed with antibodies to total polyUb, Lys63-polyUb19, Lys48-polyUb19, or unanchored Ub. Actin-normalized total-lane staining relative to control levels is presented in (b). (c) Wild-type yeast expressing MBP-Vx3, NLS-MBP-Vx3, or empty-vector controls were plated in limiting dilutions on SC-Leu medium alone or with the indicated drug. (d) HeLa cells transfected with NLS-Vx3-EGFP (green) or a non-binding control protein Vx3NB-mEGFP (green) were exposed to ionizing radiation (IR) and at intervals afterwards were fixed and stained for γH2AX (magenta) and counterstained with DAPI (blue). A subset of γH2AX foci co-stain with NLS-Vx3-EGFP (white arrows); scale bars, 10 μm. (e) For cells treated as in (d), the median number of γH2AX foci per cell are presented for untransfected control (red squares), Vx3NB-mEGFP (orange diamonds), and NLS-Vx3-EGFP (blue circles) transfected cells. The bars show the 95%-confidence limits; γH2AX foci counts in NLS-Vx3-EGFP-expressing cells versus the Vx3NB-mEGFP or untransfected controls had a p-value < 0.0001 (n ≥ 340). (f) Rap80−/− MEFs stably expressing mCherry-Rap80, transiently transfected with NLS-Vx3-EGFP and exposed to ionizing radiation, were imaged after 30 min of recovery. Scale bars, 10 μm. See also Supplementary Fig. 4.
To determine the linkage-type dependence of the cytosolic aggregates that Vx3K0-and Rx3(A7)-mEGFP bind in vivo, we used yeast strains previously engineered to express Ub from a single transgene5. We compared Vx3K0-EGFP localization in single-Ub (SU) yeast expressing wild-type Ub (SU-wt) or a Lys63R-mutant Ub (SU-Lys63R) that can produce all polyUb topologies except Lys63-linked6. Similar to the pattern in human cells, Vx3K0-EGFP localized to large, mostly perivacuolar structures in SU-wt but not SU-Lys63R yeast (Fig. 2g–i). The binding-deficient Vx3NB-EGFP was diffuse in both strains, as expected (Supplementary Fig. 2). Vx3K0-EGFP expressed in a wild-type yeast (not SU) was localized as in the SU-wt strain, indicating that this pattern was not an artifact of the strain manipulation used to replace Ub.
Next we used Vx3-EGFP to track Lys63-polyUb signals in live cells. The E3 Ub-ligase Parkin translocates to damaged mitochondria where it promotes Lys63-polyubiquitination of multiple substrates20,21. The damaged organelles are ultimately condensed into perinuclear aggregates for processing by autophagic degradation22. We transfected YFP-Parkin alone or Vx3K0-EGFP with unlabeled Parkin into HeLa cells stably expressing the mitochondrial marker mito-dsRed, and treated the cells to depolarize the mitochondria. Consistent with previous reports, YFP-Parkin rapidly relocalized to damaged mitochondria (Fig. 3a); this was followed by recruitment of Vx3K0-EGFP, presumably by Lys63-polyUb-conjugated mitochondrial proteins (Fig. 3b,c). This activity required the Ub-binding residues of Vx3 and, as HeLa cells do not express the ligase, an active, exogenous Parkin (Supplementary Fig. 3). These results indicate that our designed proteins can localize Lys63-polyUb-modified conjugates dynamically inside cells. Note that clustering of damaged mitochondria was somewhat reduced in cells also expressing Vx3K0-EGFP (Fig. 3a,b). As clustering is thought to be promoted by binding of p62 to Lys63-polyUb at the mitochondrion21, this suggests that the Lys63-polyUb was masked by bound Vx3. However, because the extent of clustering can vary with the expression levels of Vx3K0-EGFP or Parkin, this mechanism, though likely, is speculative.
Figure 3. Vx3K0-EGFP is recruited to mitochondria following uncoupling and the translocation of the E3 Ub-ligase Parkin.

(a) A representative HeLa cell transfected with YFP-Parkin (green) and mitochondria-targeted dsRed (magenta). (b) A representative HeLa cell transfected with Vx3K0-EGFP (green) along with mitochondria-targeted dsRed (magenta) and untagged Parkin. Cells were treated with 10 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) to depolarize the mitochondria and imaged every 2.5 min for 3 h. Representative images at 0, 1, and 3 h are shown. Scale bars, 10 μm. (c) Normalized average fluorescence intensities of Vx3K0-EGFP and YFP-Parkin on mitochondria over the course of CCCP treatment. Averages represent 12 (Vx3K0-EGFP) or 15 (YFP-Parkin) cells from two or more independent experiments; error bars represents. e.m. See also Supplementary Fig. 3.
PolyUb-sensor proteins as inhibitors of cellular signaling
Ub recognition by natural receptors usually is limited to a small set of residues3 and thus typically excludes simultaneous binding by other receptors or deubiquitinating enzymes. For this reason, we expected that expression of Lys63-specific polyUb sensors may inhibit Lys63-dependent signaling and specifically prevent deubiquitination of these conjugates (Supplementary Fig. 4). Indeed, expression of Rx3(A7)-mEGFP in HeLa cells for 20 h doubled Lys63-conjugates as measured by quantitative immunoblot (Fig. 4a,b). Total polyUb, Lys48-polyUb, and unconjugated Ub were largely unchanged compared to control-transfected cells, though the polyUb species appeared partially shifted to higher molecular weight forms. These results are consistent with the in vitro linkage specificities of the polyUb sensor proteins, and indicate that these constructs may specifically inhibit the action of endogenous Lys63-polyUb effectors in cells.
To test this, we expressed Vx3 fused to maltose binding protein (MBP-Vx3) in wild-type yeast on high- or low-copy plasmids (Fig. 4c). We compared these strains for their resistance to the arginine analogue canavanine or the genotoxic alkylating agent methyl methanesulfonate (MMS), previously-described phenotypes attributed to the capacity to make Lys63-polyUb6, 23. Indeed, yeast expressing MBP-Vx3 variants from high-copy plasmids were nearly as sensitive to canavanine as the SU-Lys63R strain (Fig. 4c). We did not identify the impaired function that results in canavanine sensitivity in Vx3-expressing cells, although prominent possibilities include defective arginine permease downregulation and impaired turnover of misfolded canavanine-containing proteins. Next we examined yeast growth in the presence of MMS, where Lys63-polyUb-modified proliferating cell nuclear antigen (PCNA) at sites of MMS-caused DNA damage is required to recruit specialized damage-tolerance machinery9. Remarkably, MBP-Vx3 expression restricted growth on MMS-containing plates in a dose- and compartment-specific manner. Yeast expressing NLS-MBP-Vx3 from a low-copy CEN plasmid showed inhibited growth in the presence of MMS, and yeast expressing NLS-MBP-Vx3 from the high-copy 2μ plasmid were as sensitive to MMS as the SU-Lys63R yeast (Fig. 4c). Constructs lacking the NLS did not accumulate in the nucleus (as assessed using an EGFP fusion) and had no effect.
We also examined the effect of NLS-Vx3-EGFP expression on DNA double-strand break (DSB) repair in mammalian cells, where Lys63-polyUb similarly recruits repair factors to damage sites. Expression of NLS-Vx3-EGFP in ionizing radiation (IR)-treated cells, while not affecting formation of γH2AX-containing foci, severely delayed foci disappearance (Fig. 4d,e). This is consistent with the proposed role for Lys63-polyUb downstream of the γH2AX modification at DSBs10, but upstream of repair factors important for clearance like Rap80 and BRCA110, and suggests that NLS-Vx3-EGFP inhibits DSB processing at these sites by blocking access of Rap80 and other repair factors to Lys63-modified substrates. Indeed, in response to IR, Rap80-positive foci were reduced in cells expressing NLS-Vx3-EGFP as compared to cells expressing the Vx3NB-EGFP negative control (median mCherry-Rap80-foci per cell were 5 and 19, respectively; p-value < 0.0001, n = 60) (Fig. 4f). These results establish that our designed proteins can inhibit Lys63-polyUb signaling in cellulo, and suggest that they act by displacing endogenous receptors of polyUb signals.
Resolving roles for linkage-specific polyUb in NF-κB activation
To address linkage specificity and inhibitory activity in cells, we sought a functional assay in which inhibitor expression levels could be linked to related outcomes with known Lys63-and non-Lys63-polyUb dependences. Ligand-mediated NF-κB activation is an ideal test case, since recent reports have illuminated the divergent polyUb landscape downstream of two important cell-surface receptors, interleukin-1 receptor (IL-1R) and tumor necrosis factor receptor (TNFR).
Both IL-1R and TNFR signaling follow the canonical NF-κB activation pathway (namely, ligand binding to a receptor, inhibitor of kappa B kinase (IKK) activation, and nuclear translocation of the NF-κB transcription factor)13,24. Non-degradative polyUb chains formed on one or more substrates (for example, kinase receptor interacting protein-1, RIP-1) near the receptor serve as a signaling platform to recruit a polyUb-binding subunit of IKK for activation. Originally, these chains were thought to be Lys63-linked for both TNFR and IL-1R signaling. However, a Ub-replacement strategy7 in the human U2OS osteosarcoma cell line showed that, when the sole source of Ub was Ub(Lys63Arg), IL-1β-mediated IKK activation was completely abrogated, whereas activation in the TNF-α-stimulated pathway was unaltered. More recently, mass spectrometry and linkage-specific immunoblot analyses indicated that Lys63- as well as Lys11-linked chains are present at the TNFR in roughly equal proportions after treatment with TNF-α25. The apparent resolution of these two studies is that IL-1β signaling is strictly dependent on Lys63-polyUb whereas signaling through the TNFR can function with Lys11-polyUb alone, although the typical polyUb complement is a poorly-defined mix of Lys11 and Lys63 linkages. Thus, a stringent test of our inhibitor proteins is the ability to discriminate these distinct polyUb dependencies in otherwise highly similar pathways.
We examined IL-1β- and TNF-α-stimulated NF-κB activation in U2OS cells in the presence of retrovirally-transduced Rx3(A7)-mEGFP or, as a negative-control, Vx3NB-mEGFP. We tracked activation status at the single-cell level by immunofluorescence detection of the endogenous NF-κB subunit p65, which translocates from the cytosol to the nucleus upon activation by either ligand (see Online Methods). As expected, p65 was cytosolic (translocation score 0) in unstimulated cells, but nuclear (translocation score 1) in IL-1β– or TNF-αtreated cells after 30 min (Fig. 5b). Remarkably, Rx3(A7)-mEGFP inhibited NF-κB activation in IL-1β-stimulated cells at all expression levels (Fig. 5a,b). At the lowest measurable mEGFP concentrations, inhibition was incomplete, but rapidly reached saturation upon higher Rx3(A7)-mEGFP expression, consistent with high affinity of the Rx3(A7)· Lys63-polyUb interaction (Table 1). In contrast, activation in TNF-α-treated cells was unaffected by low doses of Rx3(A7)-mEGFP (Fig. 5a,b) and, compared with stimulation by IL-1β, required ~10-fold more inhibitor (as measured by mEGFP intensity) to prevent p65 translocation. At low expression levels, where we expect the inhibitor proteins to be most specific, Rx3(A7)-mEGFP clearly distinguishes the linkage-type dependences of these two ligand-mediated NF-κB pathways and thus functions as a linkage-specific inhibitor of cellular polyUb signaling. We also found that inhibition by Vx3K0-mEGFP could distinguish IL-1β and TNF-α signaling in U20S cells, though with a somewhat smaller window of inhibitor concentrations over which linkage-specificity is observed (Supplementary Fig. 5).
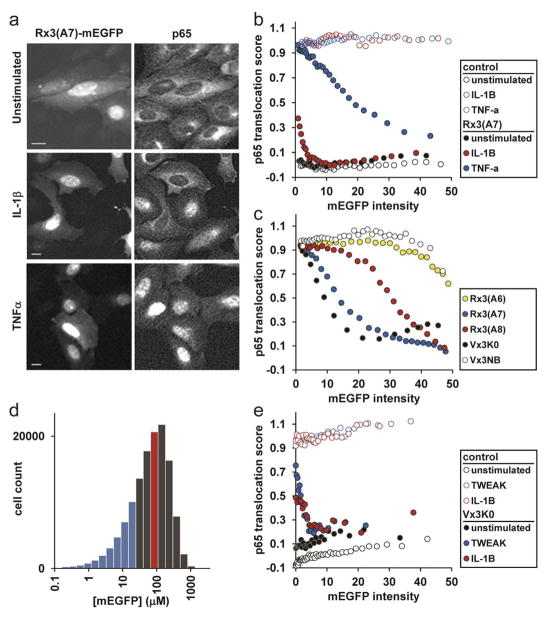
Figure 5. Differential inhibition of NF-κB activation reveals distinct roles for Lys63-linked polyUb in diverse ligand-dependent signaling pathways.
(a) U2OS cells expressing Rx3(A7)-mEGFP and stained for p65 were imaged to determine NF-κB pathway activation status after ligand treatment. Scale bars, 10 μm. (b) p65 translocation scores are plotted versus inhibitor-fusion mEGFP intensity. Each point on the plot represents the average of 500 single-cell measurements. (c) HeLa cells transiently transfected with the indicated plasmids were treated with TNF-α and scored for NF-κB activation. Each point represents the average of 1,500 single-cell measurements. (d) As in (c), HeLa cells were transiently transfected with Rx3(A7)-mEGFP. A histogram of cellular Rx3(A7)-mEGFP concentrations is shown, with the population-average level ([Rx3(A7)-mEGFP] = 87 μM) indicated by the red line and the zone of highest Lys63-linkage specificity (i.e, below the TNF-α IC50) shown in blue. (e) p65 translocation scores for retrovirally-transduced MEF cells with and without ligand treatment. Each point represents the average of 500 single-cell measurements. See also Supplementary Figs. 5 and 6.
To explore the relationship between in vitro linkage specificities and inhibitor activity in cells, we evaluated the effects of each of our tUIM-mEGFP fusions on TNF-α signaling in HeLa cells. Consistent with its higher in vitro Lys63-linkage specificity, higher cellular concentrations of Rx3(A7) were required to inhibit activation as compared to Vx3K0-mEGFP (Fig. 5c; note also the similarity to the Rx3(A7) profile in 5b); we suspect that, consistent with its lower linkage specificity in vitro, Vx3K0 is a slightly better binder for the non-Lys63-polyUb species involved in TNFR signaling. Rx3(A8)-mEGFP showed greatly reduced inhibitory activity, and Rx3(A6)-mEGFP was virtually indistinguishable from the Vx3NB non-binding control (Fig. 5c). These data taken together further suggest that the inhibition observed is due to binding to specific polyUb topologies.
These inhibitor profiles highlight the requirement to account for polyUb sensor expression levels in order to distinguish Lys63-dependent from non-Lys63-dependent signaling; with very high Rx3(A7)-mEGFP expression, both IL-1β and TNF-α signaling were completely inhibited, presumably because of weak “off-target” interactions with non-Lys63-polyUb chains such as homogeneous Lys11-polyUb, or perhaps a mixed Lys63/Lys11-polyUb species. To establish an upper limit of cellular inhibitor expression levels for linkage-specific action, we determined the IC50 of TNF-α inhibition by Rx3(A7)-mEGFP in HeLa cells by calibrating image data to quantitative immunoblots. With TNF-α signaling inhibition as a marker for non-Lys63 binding, we determined that Rx3(A7)-mEGFP above 16 μM no longer showed strong Lys63-linkage preference (Supplementary Fig. 6). This concentration agrees closely with our in vitro polyUb binding constants for non-Lys63-linked chains (Table 1), particularly when considering that effective intracellular inhibitor concentrations must be slightly lower than this because some Rx3(A7)-mEGFP typically is in perinuclear aggregates. By our estimate, the level required for linkage-specific inhibition is well below expression levels achievable from typical transient transfection of cultured cells (Fig. 5d); therefore, care must taken when applying these reagents to either account for cellular inhibitor levels explicitly at the single-cell level or, by adjusting average inhibitor expression levels (for example, through optimized transfection or a tunable induction strategy), to be within the linkage-specific concentration regime of the inhibitor.
Next, we examined signaling through the TNF-related weak inducer of apoptosis (TWEAK) receptor FN14. TWEAK ligand is a physiological activator of NF-κB that acts through signaling mechanisms that are largely unknown with respect to polyUb26. Interestingly, the ubiquitination enzymes UbcH5 and c-IAP1 are important downstream components of both TWEAK- and TNF-α-mediated NF-κB activation25, 27. To determine whether these pathways share a common polyUb-linkage dependence, we retrovirally-transduced inhibitor and control constructs into mouse embryo fibroblasts (MEFs), a cell-type that gives robust and reproducible activation of NF-κB by TWEAK. TWEAK-receptor signaling was inhibited at very low Vx3K0-mEGFP concentrations, identical to the inhibition of the strictly Lys63-dependent IL-1β signaling in both MEF and U2OS cells (Fig. 5e; compare to the IL-1β inhibition profile in 5b). We infer from this assay that TWEAK-mediated NF-κB activation shows the same strict requirement for Lys63-polyUb as the IL-1β pathway. These results show that our linkage-specific proteins may be applied across cell types to diagnose polyUb linkage-type dependences of cellular processes.
DISCUSSION
Relative to a single Ub binding domain (UBD), arrays of UBDs can exhibit enhanced affinities for polyUb; however, such high-affinity polyUb binding proteins are not necessarily linkage-specific28. Here we applied the principles of linkage-specific avidity to produce highly linkage-specific polyUb-sensor proteins from tUIMs with helical 7-amino acid linkers. tUIM linkers of lengths other than used here and other intrinsic structures may direct binding to yet other polyUb topologies; the natural complement of human tUIM linkers includes a wide variety of lengths and apparent structures, including 2-amino acid linkers that confer Lys48-polyUb selectivity14. Moreover, although the tUIM format is the most straightforward for creating avidity-based polyUb sensors, inserting UBDs into globular proteins or oligomeric protein scaffolds may provide a wider range of conformations from which to select affinity reagents for other polyUb structures15. We demonstrated here that multi-UBD constructs may be manipulated at two levels to create linkage-specific polyUb affinity reagents: individual UBD–Ub binding primarily influences overall affinity, whereas the orientation and rigidity of multi-UBD arrangements influence linkage-specificity as well. As topological complexity is increasingly revealed for other polymeric ubiquitin-like (UBL) proteins such as polySUMO4, this avidity-based design strategy similarly may be applied to create a wide range of polyUb and polyUBL affinity reagents. Antibodies are limited to the recognition of small epitopes, so it is difficult to envision a single antibody reagent targeted to a mixed polymer such as (Ub)-Lys63-(Ub)-Lys11-(Ub) or (Ub)-(SUMO)-(Ub). In contrast, our avidity-based design strategy theoretically can produce affinity reagents to complex polymer species by directing simultaneous interactions across multiple Ub or UBL linkages. We note also the potential for these reagents (and the avidity-based design strategy) to inform the search for and application of small-molecule inhibitors of specific polyUb topologies29.
Linkage-specific polyUb antibodies are increasingly available and have proven to be quite useful18,19,30. However, the requirement for disulfide-bond formation for proper folding and function of typical immunoglobulins limits those tools to mostly in vitro applications. We anticipate optimization of our probes for live-cell applications, including the possibility of FRET-based sensors that exploit conformational shifts expected for some tUIM peptides upon polyUb binding14. As intracellular inhibitors, the contributions of subcellular localization to polyUb signaling may be explored by appending targeting sequences to the protein fusions. Finally, because the sensor proteins inhibit deubiquitination (Fig. 4a,b), their expression in cells may allow the accumulation of otherwise unstable polyUb conjugates to levels sufficient for isolation and identification. However, as our study shows, sensor-protein concentration must be carefully controlled to avoid off-target binding that is increasingly favored above the dissociation constant for Lys63-polyUb (Fig. 5d). Unlike Ub-replacement strategies in mammalian cells that require the coordinated inducible expression and knockdown of multiple Ub genes7, our inhibitor strategy is easily applied to any cell line, tissue-type, or organism that can be transfected or virally transduced. Moreover, this type of study should be extensible (for example, via microscopy or FACS) to any cellular function with a scorable single-cell phenotype. Recent studies of TNFR-mediated signaling underscore key differences in the available approaches to study linkage-specific polyUb functions. Ub-replacement strategies restrict which linkages can be made and thus can reveal a specific Ub–Ub linkage requirement2,6, but no effect will be observed if an alternative linkage-type can serve an equivalent signaling role. Conversely, mass spectrometry and linkage-specific immunoblot approaches reveal which linkages are present25, but give no information about their relative importance for function. Our linkage-specific inhibitor strategy combines useful aspects of both approaches by linking competitive inhibition of naturally-assembled polyUb signals to a functional outcome. Lastly, we note that, unlike Ub-replacement strategies used to manipulate Ub pools in vivo, expression of polyUb-sensor proteins causes dose-sensitive and thus (most likely) reversible Lys63-polyUb-associated phenotypes. This makes possible genetic screen approaches to identify function-specific Lys63-polyUb receptors, whose overexpression may restore function by moving receptors into concentration regimes where they can effectively compete with inhibitors for Lys63-polyUb binding.
ONLINE METHODS
Plasmids and proteins
We created designed tUIM genes by overlapping primer-extension PCR with 20–40 base oligonucleotides. Cloning and sequence details are summarized in Supplementary Tables 1 and 2. mEGFP used in fusion proteins was described previously31. For binding assays, we bacterially expressed His6-tagged tUIMs and purified them using Ni-NTA agarose (Qiagen) according to the manufacturer’s directions. We labeled recombinant tUIMs on a single cysteine residue with fluorescein maleimide or biotin maleimide (Pierce), and further purified them by gel filtration or anion-exchange chromatography as needed. We produced polyUb chains of defined length and linkage as previously described32, 33.
Polyubiquitin binding assays
We performed direct fluorescence anisotropy binding titrations as described14, with fluorescently-labeled peptide present at 1, 4, 10, or 100 nM in the cuvette, depending on the anticipated affinity of the peptide being tested. We made binding measurements at 25 °C in fluorescence buffer (25 mM Na phosphate pH 7.4,150 mM NaCl, 5 mM β-mercaptoethanol, 1 mM EDTA, and 0.05% Brij35). To reduce the amount of polyUb needed to determine Kd values for weakly-binding proteins, we performed some titrations in a competition assay format (Supplementary Fig. 1). Typically, we combined fluorescein-labeled binding peptide (e.g., 4 nM fluorescein-Vx3) with polyUb (e.g., 50 nM Lys63-Ub3) to give ~80% saturation; we determined fluorescence anisotropy values with various amounts of unlabelled competitor protein and fit Kd values with a model of competitive binding to a single site; we derrived baseline anisotropy values from samples with no competitor protein, and the plateau value from fluorescent binding protein without polyUb ligand. Where relatively weak binding was measured, high protein concentrations (> 10 μM) caused anisotropy values to increase slightly due to changes in the solution viscosity. In those cases, we determined corrections for the viscosity effects by titrations of fluorescent binding peptide with competitor protein in the absence of polyUb. For the far-western dot blot, we spotted the indicated amounts of purified ubiquitin species in binding buffer plus 1 mg/ml BSA directly onto dry PVDF membrane and allowed them to dry completely. We blocked the methanol-wetted membranes in 3% BSA, probed the membranes with 30 nM biotinylated His6-Vx3 peptide, washed in TST, probed with streptavidin-HRP (Pierce), and washed again with TST; detection was by chemiluminesence. Antibodies with specificity to total polyUb were from Millipore (clone FK2) and antibodies to unanchored Ub were from Epitomics (Ub C-terminus antibody; clone RPS27A).
Yeast and mammalian cell culture
We cloned MBP-Vx3 and NLS-MBP-Vx3 into pRS415-ADH (low-copy CEN plasmid) or pRS425-ADH (high-copy 2μ plasmid), each containing the constitutive alcohol dehydrogenase (ADH) promoter. We transformed these into wild-type yeast strain BY4741 (MATa leu2Δ0 his3Δ1 ura3Δ0 met15Δ0) and transformants were selected on SC-Leu medium. In the original (SUB280-derived) single-Ub yeast strains, LEU2 was used to replace UBI4. To recover the leu2 marker in these strains, we replaced via homologous recombination the LEU2 gene at the UBI4 locus with a cassette that conferred hygromycin resistance. We selected hygromycin-resistant leucine auxotrophs, and confirmed the proper integration by PCR. For yeast growth assays, we grew two independent transformants from each transformation to mid-log phase in SC-Leu medium and spotted 0.25 OD600 equivalents (~3×106 cells) in serial 10-fold dilutions on the following plates: SC-Leu (control), SC-Leu containing 0.025% methyl methanesulfonate (MMS), or SC-Leu-Arg containing 0.0001% canavanine.
We maintained HeLa and MEF cell lines in DMEM, and U2OS cells were grown in McCoy’s medium, both media supplemented with 10% fetal bovine serum, 100 I.U. penicillin, and 100 μg/ml streptomycin. We introduced mCherry-Rap80 into Rap80−/− MEFs (a kind gift from J. Chen, University of Texas M.D. Anderson Cancer Center, Houston, TX) by retroviral transduction, followed by IL-2Rα-mediated bead-sorting to select a population of cells that stably express the construct. We introduced polyUb sensor constructs by lipid transfection using Lipofectamine 2000 (Invitrogen) or Fugene6 (Roche), or by retroviral transduction using the RetroX packaging cell line (Clontech). Confluent HeLa cells in 6-well dishes transfected with 1 μg of inhibitor or control plasmids were used for ubiquitin immunoblot analysis. We lysed cells on ice for 30 min in PBS with 1% Triton X-100, 20 mM NEM, 10 mM EDTA, and mini-Complete protease inhibitor cocktail (Roche). Antibodies in Fig. 4a were used according to the manufacturer’s instructions, and immunoblots were quantified using IR800 secondary antibodies (LI-COR).
Microscopy
For imaging Vx3-EGFP in the single-Ub yeast strains, we cloned Vx3K0-EGFP and NLS-Vx3K0-EGFP into pRS425-ADH, transformed them into yeast strains BY4741 or SUB2806 (expressing wild-type or Lys63Arg Ub), and selected transformants on SC-Leu. We fixed mid-log phase cells for 1 h with 3.7% formaldehyde, washed with PBS, and then examined using a 100x objective. For HeLa and MEF imaging, we fixed cells 18 h after transfection with 2% paraformaldehyde for 10 min, mounted them using ProLong Gold antifade medium (Invitrogen) with DAPI, and imaged the cells with a 60x objective and deconvolution. We transiently transfected Rap80−/− MEF cells expressing mCherry-Rap80 with NLS-Vx3K0-EGFP or Vx3NB-EGFP by using Fugene6 reagent. After 24 h, we exposed cells to 3 Gy of γ-irradiation and allowed them to recover for 30 min at 37 °C before fixation with 2.5% paraformaldheyde. We mounted cells on slides using ProLong Gold medium and aqcuired fluorescence images with an Olympus IX81 spinning-disk confocal microscope (CSU22 head) using a 60x/NA 1.42 oil objective. For the γH2AX foci counts, we irradiated HeLa cells (1 Gy) 24 h after transient transfection. After the indicated recovery times, we fixed cells in 2.5% paraformaldehyde for 10 min, permeabilized them in 0.1% Triton X-100, stained them with anti-phospho-histone H2A.X (Ser139) (clone JBW301, Upstate) for 2 h at 1:100 dilution, incubated them with Alexa Fluor 594 anti-mouse IgG (1:300 dilution), counterstained nuclei with DAPI, and mounted the coverslips on slides using ProLong Gold. Statistical analyses of foci counts employed the non-parametric Wilcoxon rank-sum test. For mitophagy experiments, we imaged live cells in CO2-independent media (Invitrogen) using an UltraView LCI confocal microscope (PerkinElmer) at 37 °C with a 63x/NA1.4 Apochrome objective. We used Volocity software (PerkinElmer) for analysis. We calculated the fluorescence intensity of the YFP or GFP-tagged proteins in the area of the mitochondria, normalized this to the total cell YFP or GFP, and set the start value to one. For fixed-cell mitophagy controls (Supplementary Fig. 3), we plated HeLa cells onto chambered coverglasses where they were cultured overnight. We then transiently transfected the cells with Vx3K0-EGFP or Vx3NB-EGFP plasmids. The next day, we treated cells with 10 μM CCCP for 3 h, then fixed and stained them for the mitochondrial marker Tom20 (anti-Tom20 polyclonal antibody; Santa Cruz, sc-11415). We collected fixed-cell images on a Zeiss LSM 510 confocal microscope using a 63x/NA 1.40 Apochrome objective. We adapted the staining protocol for LC3 (Supplementary Fig. 2) from a method kindly provided by J. Gutierrez (Millennium – Takeda Pharmaceuticals). 24 h after transfection (Lipofectamine 2000) we fixed cells with 2.5% paraformaldehyde for 10 min, permeabilized them with 100 μg/ml digitonin for 15 min, and blocked for 1 h with 0.5% blocking reagent (Roche #11096176001) in 100 mM Tris pH 7.5, 150 mM NaCl. Subsequently, we incubated the cells with anti-LC3 antibody (clone 4E12, MBL International; 1:300 dilution) overnight at 4 °C, stained the sample with Alexa Fluor 594 anti-mouse IgG, and mounted the coverslips on slides using ProLong Gold medium. We obtained fluorescence images with the Olympus IX81 spinning-disk confocal microscope using a 100x/NA1.4 oil objective.
NF-κB activation experiments
We measured NF-κB activation at the single-cell level by fluorescence microscopy34. We plated U2OS (5,000 cells), HeLa (9,000 cells), or MEF (5,000 cells) cells in clear-bottom 96-well imaging plates. After overnight growth, we transfected or retrovirally transduced the cells as described above; we changed the medium 18–24 h after transfection or transduction and 1 h prior to the ligand stimulation. We added TNF-α (PeproTech) or IL-1β (Pierce) to the growth medium at 10 ng/ml, and TWEAK (PeproTech) 100 ng/ml. All doses were the minimum that gave reproducible p65 translocation after 30 min. We chose the 30 min time point to avoid the potential complications of apoptosis and autocrine signaling from NF-κB transcriptional targets, both of which have been shown to occur at longer times35,36. After stimulation, we fixed cells as above, permeabilized them in PBS with 0.2% Triton X-100 for 10 min, blocked with LI-COR Blocking Buffer, incubated with anti-p65 monoclonal antibody (F-6, Santa Cruz) diluted 1:300 in blocking buffer, washed, and stained the plates with Alexa Fluor 647-conjugated anti-mouse IgG (Invitrogen) diluted 1:2000 in blocking buffer. We stained DNA and total protein with a combination of Hoechst 33342 (Invitrogen) and Whole Cell Stain Blue (Pierce). Images were collected at 10x magnification by automated microscopy (ArrayWorx, API).
We used the custom image analysis software Imagerail34 to define cell and nuclear boundaries from empirical threshold values determined on the blue channel. After segmentation, we gated out untransfected cells from the analysis based on the EGFP intensity. We removed the small number of deformed, dead, or incorrectly segmented cells from the analysis by gating on features of cell size, nuclear size, and the ratio of nuclear-to-cytoplasmic intensities on the blue and green channels. Finally, we measured p65 translocation as the ratio of nuclear-to-cytoplasmic staining for each cell on the Cy5 (i.e., Alexa Fluor 647) channel. Measuring the disappearance of IκBα at earlier time points (15 and 20 min), while less robust, gave very similar results with respect to TWEAK and IL-1β inhibition (data not shown). We assigned a p65 translocation score of 0 to the average nuclear/cytoplasmic p65 ratio for unstimulated control cells (Vx3NB-mEGFP), and 1 for the control cells treated with ligand. Untransfected and control-transfected cells gave very similar responses in terms of p65 translocation. We then scaled the nuclear/cytoplasmic ratio for cells transfected with inhibitor proteins between 0 and 1 based on the control-cell analysis.
Control experiments established the equivalence of Vx3NB-EGFP and EGFP alone as negative controls and of Vx3-EGFP and Vx3K0-EGFP as inhibitors (Supplementary Fig. 5). At most transfection levels, the negative control Vx3NB-mEGFP had little effect on the NF-κB-activation status of unstimulated cells; however, we note that some constitutive NF-κB activation was apparent at very high transfection levels of all plasmids. This constitutive activation seemed worse for transient transfection than for retroviral transduction, and worse in U20S cells than in HeLa and MEF cells, but the effect was somewhat variable from experiment to experiment depending on transfection conditions, the precise time between transfection and fixation, and the inhibitor expression level.
Supplementary Material
Supplementary Figure 1. Additional structural and binding data for designed tUIM peptides.
Supplementary Figure 2. Subcellular localization patterns of designed tUIM fluorescent fusion proteins expressed in cultured human cells.
Supplementary Figure 3. HA-Vx3K0-GFP is recruited to mitochondria in the presence of Parkin whereas HA-Vx3NB-GFP is not.
Supplementary Figure 4. Vx3-fusions inhibit polyUb recognition and cleavage.
Supplementary Figure 5. Control experiments for using fluorescent fusions of polyUb-sensor peptides as linkage-specific inhibitors inside cells.
Supplementary Figure 6. Calculating the IC50 of TNF-α inhibition by expressed Rx3(A7)-mEGFP.
Supplementary Table 1. Cloning details for all constructs used in this study.
Supplementary Table 2. Sequences of all tUIM peptides used in this study.
Acknowledgments
We thank S. Beese-Sims for constructing the leucine-auxotrophic version of the single-ubiquitin yeast strains, P. Sorger and B. Millard for helpful discussions about the manuscript and technical assistance on NF-κB activation experiments, A. Sliva for assistance with the yeast experiments, B. Schmitt for assistance with binding experiments, A. Hess for help with statistical analyses, and T. Yao for many helpful discussions. J.J.S. is a fellow of the Damon Runyon Cancer Research Foundation (DRG#2073-11). Work in this study was supported, in part, by NIH Common Fund grant RR020839 (J.D.B.), NIH grant P01 CA139980 (P. Sorger), NINDS Intramural Program (R.J.Y.), and NIH grants RC1 GM091424 (R.E.C.) and 1R01 GM097452 (R.E.C.).
Footnotes
AUTHOR CONTRIBUTIONS
J.J.S. developed the tUIM design strategy; J.J.S. and R.E.C. conceived of the study; J.J.S., E.M.C., F.S., L.A.K., and R.E.C. performed the experiments; all authors contributed to experimental design and data analysis; J.J.S. and R.E.C. wrote the manuscript with input from all of the authors.
COMPETING FINANCIAL INTERESTS
A U.S. patent application describing design and applications of linkage-specific polyubiquitin binding proteins has been filed by Johns Hopkins University on behalf of J.J.S. and R.E.C.
References
- 1.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains -from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finley D, et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 9.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 10.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 12.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010;20:196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat Struct Mol Biol. 2009;16:883–889. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser SE, et al. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitinpools. Nat Methods. 2011 doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto ML, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 20.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 21.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnason T, Ellison MJ. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 25.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh T, et al. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J Biol Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 27.Feltham R, et al. Tumor necrosis factor (TNF) signaling, but not TWEAK (TNF-like weak inducer of apoptosis)-triggered cIAP1 (cellular inhibitor of apoptosis protein 1) degradation, requires cIAP1 RING dimerization and E2 binding. J Biol Chem. 2010;285:17525–17536. doi: 10.1074/jbc.M109.087635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hjerpe R, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma R, et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–120. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, et al. Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin. Proc Natl Acad Sci U S A. 2008;105:20197–20202. doi: 10.1073/pnas.0810461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 32.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 33.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millard BL, Niepel M, Menden MP, Muhlich JL, Sorger PK. Adaptive informatics for multifactorial and high-content biological data. Nat Methods. 2011;8:487–492. doi: 10.1038/nmeth.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janes KA, et al. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 36.Janes KA, et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Additional structural and binding data for designed tUIM peptides.
Supplementary Figure 2. Subcellular localization patterns of designed tUIM fluorescent fusion proteins expressed in cultured human cells.
Supplementary Figure 3. HA-Vx3K0-GFP is recruited to mitochondria in the presence of Parkin whereas HA-Vx3NB-GFP is not.
Supplementary Figure 4. Vx3-fusions inhibit polyUb recognition and cleavage.
Supplementary Figure 5. Control experiments for using fluorescent fusions of polyUb-sensor peptides as linkage-specific inhibitors inside cells.
Supplementary Figure 6. Calculating the IC50 of TNF-α inhibition by expressed Rx3(A7)-mEGFP.
Supplementary Table 1. Cloning details for all constructs used in this study.
Supplementary Table 2. Sequences of all tUIM peptides used in this study.