Abstract
Purpose
Metastasis is responsible for the death of most cancer patients, yet few therapeutic agents are available which specifically target the molecular events that lead to metastasis. We recently showed that inactivating mutations in the tumor suppressor gene BAP1 are closely associated with loss of melanocytic differentiation in uveal melanoma and metastasis (UM). The purpose of this study was to identify therapeutic agents that reverse the phenotypic effects of BAP1 loss in UM.
Experimental Design
In silico screens were performed to identify therapeutic compounds predicted to differentiate UM cells using Gene Set Enrichment Analysis and Connectivity Map databases. Valproic acid, trichostatin A, LBH-589 and suberoylanilide hydroxamic acid were evaluated for their effects on UM cells using morphologic evaluation, MTS viability assays, BrdU incorporation, flow cytometry, clonogenic assays, gene expression profiling, histone acetylation and ubiquitination assays, and a murine xenograft tumorigenicity model.
Results
HDAC inhibitors induced morphologic differentiation, cell cycle exit, and a shift to a differentiated, melanocytic gene expression profile in cultured UM cells. Valproic acid inhibited the growth of UM tumors in vivo.
Conclusions
These findings suggest that HDAC inhibitors may have therapeutic potential for inducing differentiation and prolonged dormancy of micrometastatic disease in UM.
Keywords: BAP1, differentiation, eye, HDAC inhibitor, metastasis, uveal melanoma, valproic acid
INTRODUCTION
Metastasis is responsible for the death of most cancer patients, yet there are few therapies available that effectively target the metastatic process. This is particularly true for metastatic melanoma, which is notoriously resistant to treatment. Since metastatic melanoma may remain asymptomatic in a state of dormancy for months to years prior to becoming clinically detectable, one therapeutic strategy is to prevent or delay micrometastatic disease from escaping dormancy by inducing senescence or differentiation (1). However, such a strategy has been hampered by an inadequate understanding of the genetic events driving metastasis and, consequently, a lack of rational drug targets.
Uveal melanoma (UM) is a highly aggressive form of melanoma that exhibits a strong tendency for lethal hematogenous metastasis to the liver and other sites (2). At the time the primary eye tumor is diagnosed, less than 4% of patients have detectable metastatic disease (3). Yet up to half of these individuals will eventually succumb to metastasis despite successful treatment of the primary tumor, indicating that they harbored subclinical micrometastases at initial presentation. The most accurate method for identifying UM patients who are at high risk of metastasis is by gene expression profiling of the primary tumor, which can be performed on a fine needle biopsy using a validated 15-gene assay (4). This method separates UMs into two classes. Class 1 tumors, which have a very low metastatic risk, are composed of more differentiated tumor cells, and their gene expression profile is highly similar to that of normal differentiated melanocytes (5). In contrast, class 2 tumors, which have a high risk of metastasis, contain cells that have lost morphologic features of melanocytic differentiation, and their gene expression signature is enriched for genes expressed in primitive neuroectodermal cells (5).
We recently showed that inactivating somatic mutations in the tumor suppressor gene BAP1, located at chromosome 3p21, and concomitant loss of the other copy of chromosome 3 are present in the vast majority of class 2 tumors but not class 1 tumors (6). Depletion of BAP1 in cultured class 1 UM cells induced a loss of melanocytic differentiation and acquisition of a class 2 gene expression profile, suggesting that the loss of BAP1 may be mechanistically linked to metastasis. Functional studies have confirmed the tumor suppressor activity of BAP1 (7, 8), and several recent reports have linked germline BAP1 mutations to a spectrum of familial cancers including uveal and cutaneous melanoma and mesothelioma (9–11). The link between BAP1 and differentiation in UM suggests that therapeutic compounds which trigger differentiation in UM cells may have clinical value, particularly in the adjuvant setting where the goal would be to induce a prolonged latency of micrometastatic disease. Thus, we performed two complementary in silico screens to identify therapeutic compounds predicted to shift UM cells from the class 2 to the class 1 signature. Histone deacetylase (HDAC) inhibitors were ranked at or near the top of candidate compound lists in both screens.
We analyzed the effects of four different HDAC inhibitors, including valproic acid (VPA), trichostatin A (TSA), LBH-589 and suberoylanilide hydroxamic acid (SAHA), in established UM cell lines and in primary UM cells in short term culture. These compounds induced a proliferation block through G1 cell cycle arrest, as well as morphologic and transcriptomic changes consistent with melanocytic differentiation. VPA inhibited the growth of UM tumors in vivo. We conclude that HDAC inhibitors may play a role in the adjuvant therapy of patients with UM by inducing differentiation and prolonged dormancy of micrometastatic disease.
MATERIALS AND METHODS
Tumor samples
This study was approved by the Institutional Review Board of Washington University in St. Louis and adhered to the tenets of the Declaration of Helsinki. Primary UM samples were collected at the time of enucleation as previously described (12). Informed consent was obtained for each patient. All samples were confirmed to be uveal melanomas by pathologic evaluation. Tumor samples were snap-frozen for DNA and RNA isolation or collected in HAM’S F-12 medium for tissue culture. Primary UM cells were grown in serum-free MDMF medium on collagen-covered tissue culture plates in 5% CO2 and 4% O2 (13). Elimination of contaminating cells was confirmed by microscopic inspection and Melan-A staining. The multi-gene prognostic assay for assignment of tumors to class 1 or class 2 groups was performed as described elsewhere (4). To determine BAP1 status of primary tumors, genomic DNA was subjected to Sanger sequencing of all BAP1 exons as described elsewhere (14). The UM cell lines 92.1, OCM1A and Mel202 (kindly provided by Drs. M. Jager, J. Kan-Mitchell and B. Ksander, respectively) were grown in RPMI-1640 supplemented with 10% FBS, L-glutamine and antibiotics at 5% CO2. These UM cell lines are all BAP1-wildtype with a gene expression profile similar to class 1 tumors.
Gene Set Enrichment Analysis (GSEA) and Connectivity Mapping
Gene expression profiles from fourteen class 1 and ten class 2 UMs were obtained on the Affymetrix U133A platform, as previously described (15). Curated gene sets enriched in class 1 UM tumors versus class 2 UM tumors were identified with GSEA (version 2.0.4; Broad Institute), using a significance threshold set at a nominal P-value < 0.01 (16), as we previously described (5). To identify compounds that could potentially induce differentiation of class 2 UM cells, we interrogated the Connectivity Map database (Broad Institute) (17) for compounds that caused gene expression changes that closely resembled the gene expression differences between normal uveal melanocytes versus class 2 UM cells, as well as class 1 UM cells versus class 2 UM cells. The 100 most differentially expressed genes were chosen using Student’s t-Test. This gene set was compared to those from the Connectivity Map database, and potential compounds were chosen using the following parameters (e.g., positive enrichment score with a P<0.05).
Chemosensitivity assays
The effects of four HDAC inhibitors, VPA (Sigma), TSA (Sigma), LBH-589 (Selleck) and SAHA (Selleck), were studied in UM cell lines and primary UM cells. Drug concentrations ranged from 0.5–2 mM for VPA, 50–250 nM for TSA, 5–60 nM for LBH-589 and 0.5–2.5 µM for SAHA. DMSO was added as vehicle in TSA, LBH-589 and SAHA untreated controls.
MTS assays were performed according to manufacturer’s instructions (CellTiter 96® AQueous Assay reagent; Promega). Bromodeoxyuridine (BrdU; 1:1000; Amersham Biosciences) incorporation assays were performed using an anti-BrdU-peroxidase-conjugated antibody according to the manufacturer’s instructions (Roche; dilution 1:75). Tetramethylbenzidine (TMB; Sigma) was added as substrate for signal detection and colorimetric changes were measured at 370 nm using a microplate spectrophotometer (SpectraMAX 190; Molecular Devices). Flow cytometry was performed after treatment of cells for 48h using a standard propidium iodide staining protocol as previously described (18) using a FACScan analyzer (BD Biosciences). The percentage of cells in each phase was determined with the VenturiOne software (Applied Cytometry). For clonogenic assays, flow cytometry (Moflo; Cytomation) was used to seed one viable cell per well in ultra-low attachment 96-well plates containing MDMF medium only or MDMF with drug. Cells were monitored with phase contrast microscope, and cell number was assessed for each well at 7 days. Cell morphology was assessed by phase contrast microscopy and morphologic changes were quantified by counting the number of dendritic arborizations per cell in primary UM cells or by determining the maximal cell length/width ratio (spindle morphology index) in UM cell lines using ImageJ. Higher values are consistent with increasing melanocytic differentiation.
Multi-gene prognostic assay and qPCR
Total RNA extracted with TRIzol (Invitrogen) was DNase-treated and reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA was pre-amplified for 14 cycles with pooled primers according to the manufacturer’s protocol (Applied Biosystems). The multi-gene prognostic assay was performed with RNA from primary tumor cells treated with VPA, LBH-589 and SAHA, and molecular class was determined using Support Vector Machine (SVM), as described elsewhere (4). BAP1 mRNA levels were measured by qPCR as previously described (6).
BAP1 knockdown
BAP1 was depleted in the 92.1 UM cell line by using a lentiviral-based shRNA. Lentiviral pLKO.1 shRNA vectors for GFP (clonetechGfp-438s1c1) and BAP1 (NM_004656.2-321s1c1) (developed by the RNAi Consortium (TRC)) were purchased from the Children's Discovery Institute/Genome Sequencing Center at Washington University in St. Louis. Viral production and infections were carried out according to the The RNAi Consortium recommendations (Broad Institute). Lentiviruses encoded by the pLKO.1 shRNA vectors were packaged in 293FT cells (Invitrogen) after cotransfection of the shRNA plasmids with pCMV-dR8.2 dvpr and pCMV-VSV-G lentiviral plasmids (Addgene plasmids #8454 and #8455) using TransIT-LT1 (Mirus Bio) (19). 92.1 UM cell line was infected for 24 hours with lentiviral supernatants in the presence of 8 µg/ml polybrene. Puromycin (3 µg/ml) was added to the cells at 24 hours post-infection for selection.
Western blotting and immunofluorescence
Western blotting was performed as previously described (18) using anti-BAP1 (1:250; Santa Cruz), anti-alpha-tubulin (loading control, 1:1000; Sigma), anti-acetyl-histone H3Lys9 (1:200; Cell signaling), anti-histone H3 (1:200; Cell signaling), anti-ubiquityl-histone H2A (1:100; Millipore), and anti-histone H2A (1:200; Millipore) antibodies. Histone proteins for western blotting were extracted from cells according to previously published protocol (20). Western blotting densitometry was performed with the AlphaEaseFC software using the Spot Denso Analysis Tool. Intensity of each Ub-H2A and total H2A bands was measured using a rectangular spot and Ub-H2A was normalized to total H2A after background subtraction. Immunofluorescence was performed by plating 5×103 92.1 cells on Lab-Tek 8-well Permanox chamber slides (Nunc) and using anti-BAP1, anti-acetyl-histone H3Lys9 and anti-ubiquityl-histone H2A antibodies.
Animal studies
Animal experiments were approved by the Washington University in St. Louis Animal Studies Committee. 1×106 92.1 UM cells were resuspended in Cultrex (Trevigen) and injected subcutaneously into the flanks of NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ JAX® (NOD SCID gamma) mice (Jackson Laboratory). Mice were then given intraperitoneal injection of VPA (0.1 mg per g of body weight) every other day, beginning at day 7. Tumor volume was monitored once a week and the mice were euthanized after 42 days. Tumors were collected and volume measured using the ellipsoid volume formula (π xyz/6 mm3).
Statistical analysis
Except where otherwise noted, data were analyzed for statistical significance using MedCalc software, version 9.5.1.0.
RESULTS
In silico screening for compounds that reverse the effects of BAP1 loss
BAP1 loss in UM cells results in morphologic and transcriptomic changes consistent with a loss of melanocytic differentiation and a shift from class 1 to class 2 transcriptomic profile (6). Thus, we sought to identify therapeutic compounds that may reverse the effects of BAP1 loss by restoring a more differentiated, class 1-like transcriptomic profile. We used two complementary in silico approaches – GSEA and Connectivity Mapping – to compare genes that are differentially expressed between class 1 and class 2 UMs to curated gene sets associated with perturbation of cancer cells with therapeutic compounds. Using GSEA, the gene set that was most similar to the genes up-regulated in class 1 UMs (relative to class 2) was PEART_HISTONE_UP (Fig. 1), which consisted of genes up-regulated by the HDAC inhibitors SAHA and depsipeptide (21). We obtained similar results with the Connectivity Mapping, which identified three HDAC inhibitors (VPA, TSA and SAHA) among its top matches (Supplementary Table S1).
Figure 1. Gene set enrichment analysis.
Enrichment plot of gene set PEART_HISTONE_UP. The gene set that was most similar to the genes up-regulated in class 1 UMs (relative to class 2) using GSEA was PEART_HISTONE_UP, which consisted of genes up-regulated by the HDAC inhibitors SAHA and depsipeptide. Genes that are overrepresented in class 1 tumors show a peak enrichment score (ES) that is positive and to the left of the plot, and those that are overrepresented in class 2 tumors show a peak ES that is negative and to the right of the plot.
HDAC inhibition blocks proliferation of UM cells
Initially we chose VPA to test the effects of HDAC inhibition in UM cells. As expected, VPA caused a dose-dependent increase in histone H3 acetylation (Supplementary Fig. S1). In all three UM cell lines (92.1, OCM1A and Mel202), VPA inhibited proliferation but did not significantly reduce the fraction of viable cells, induced a G1 cell cycle arrest and markedly reduced the clonogenicity of UM cells (Fig. 2). The spindle morphology index increased after treatment with the HDAC inhibitors (Supplementary Fig. S2). Similar changes were seen with TSA and LBH-589, except that these agents significantly reduced the fraction of viable cells and increased the proportion of cells undergoing apoptosis (Fig. 2), consistent with increased cytotoxicity.
Figure 2. Effects of HDAC inhibitors on UM cell lines.
Cells were either untreated (UT) or treated with VPA, TSA and LBH-589. A, MTS cell viability assays after 72 hours treatment. The absorbance of control cells at 490 nm was taken as 100%. B, BrdU incorporation assays after 72 hours treatment. The absorbance at 370 nm of control cells was taken as 100%. C, cell cycle analysis by flow cytometry using propidium iodide staining. Cells were either untreated (UT) or HDAC inhibitor-treated for 48 hours; x-axis represents DNA content and y-axis represents cell number. D, single-cell clonogenic assays. OCM1A were either untreated (UT) or HDAC inhibitor-treated and cell number for each well containing a colony was assessed after 7 days. *P<0.05.
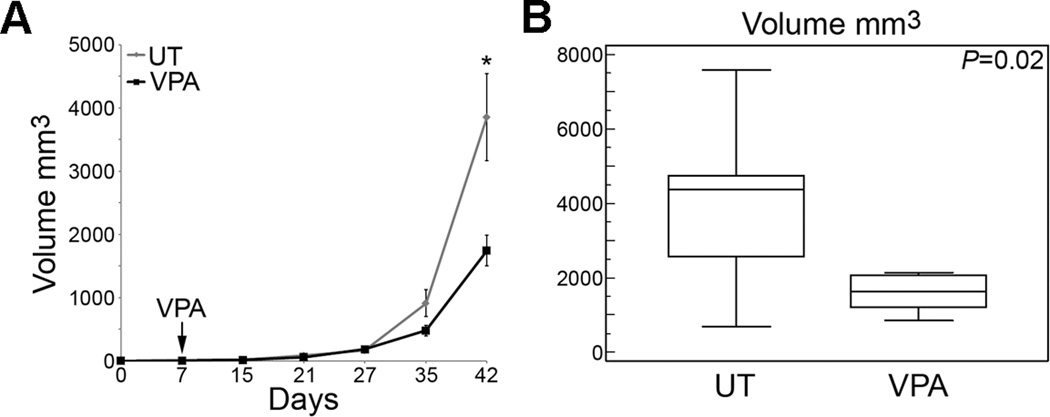
VPA inhibits UM tumor growth in vivo
Since VPA induced cell cycle arrest rather than cell death, we predicted that it may have therapeutic potential in an adjuvant setting to block the growth of micrometastatic disease. To model this situation, we established small subcutaneous flank tumors in NOD SCID gamma mice and tested whether VPA could prevent the growth of these tumors. Treatment with VPA or control was initiated when the tumors became barely palpable (usually about 7 days after injection). VPA markedly reduced the growth and final volume of these tumor deposits compared to control (Fig. 3).
Figure 3. VPA inhibits the growth of UM tumors in vivo.
A, growth curves of 92.1 cell xenografts in NOD SCID gamma mice, either untreated (UT) or treated with VPA (0.1 mg per g of body weight, injected intraperitoneally every other day, from day 7 to day 41; n=10 animals for each condition). B, volume of untreated (UT) and VPA-treated flank tumors at the time of necropsy (day 42). Middle line represents median; box, 25th to 75th percentiles; outer bars, minimum and maximum values. *P<0.05.
BAP1 loss sensitizes UM cells to HDAC inhibition
BAP1 is the catalytic subunit of the PR-DUB complex that deubiquitinates histone H2A (22). As expected, RNAi-mediated knock down of BAP1 in UM cells (Fig. 4A) induced a marked increase in H2A ubiquitination (Fig. 4B, upper panel). Since a recent report showed that HDAC inhibitors decrease histone H2A ubiquitination through transcriptional repression of the PRC1 component BMI1 (23), we wished to determine whether HDAC inhibition may reverse this H2A hyperubiquitination in BAP1-deficient UM cells. Indeed, VPA markedly reduced the levels of histone H2A ubiquitination in BAP1-depleted cells (Fig. 4B, lower panel and Fig. 4C upper panel), while total histone H2A level remained unchanged (Fig. 4C, lower panel). We showed in Fig. 2A that VPA inhibited proliferation of BAP1-wildtype UM cell lines, but it did not reduce the fraction of viable cells. However, when we stably depleted BAP1 in UM cells using lentiviral shRNA, VPA significantly reduced the fraction of viable cells compared to control cells (Fig. 4D), indicating that BAP1 loss sensitized the UM cells to HDAC inhibition.
Figure 4. VPA counteracts the abnormal ubiquitination of histone H2A in BAP1-depleted UM cells.
A, BAP1 western blot (left panel) and immunofluorescence (right panel, magnification 200X) of 92.1 control (shGFP) and BAP1-depleted (shBAP1) cells. B, ubiquityl-histone H2A (Ub-H2A) immunofluorescence of untreated (UT) or VPA-treated 92.1 control (shGFP) and BAP1-depleted (shBAP1) cells. Magnification 400X. C, ubiquityl-histone (Ub-H2A) and total histone H2A western blots (left panel) of untreated (UT) or VPA-treated 92.1 BAP1-depleted cells (shBAP1); corresponding densitometry (right panel). D, MTS cell viability assays after 72 hours treatment with VPA 2 mM. The absorbance of control cells at 490 nm was taken as 100%. *P<0.05.
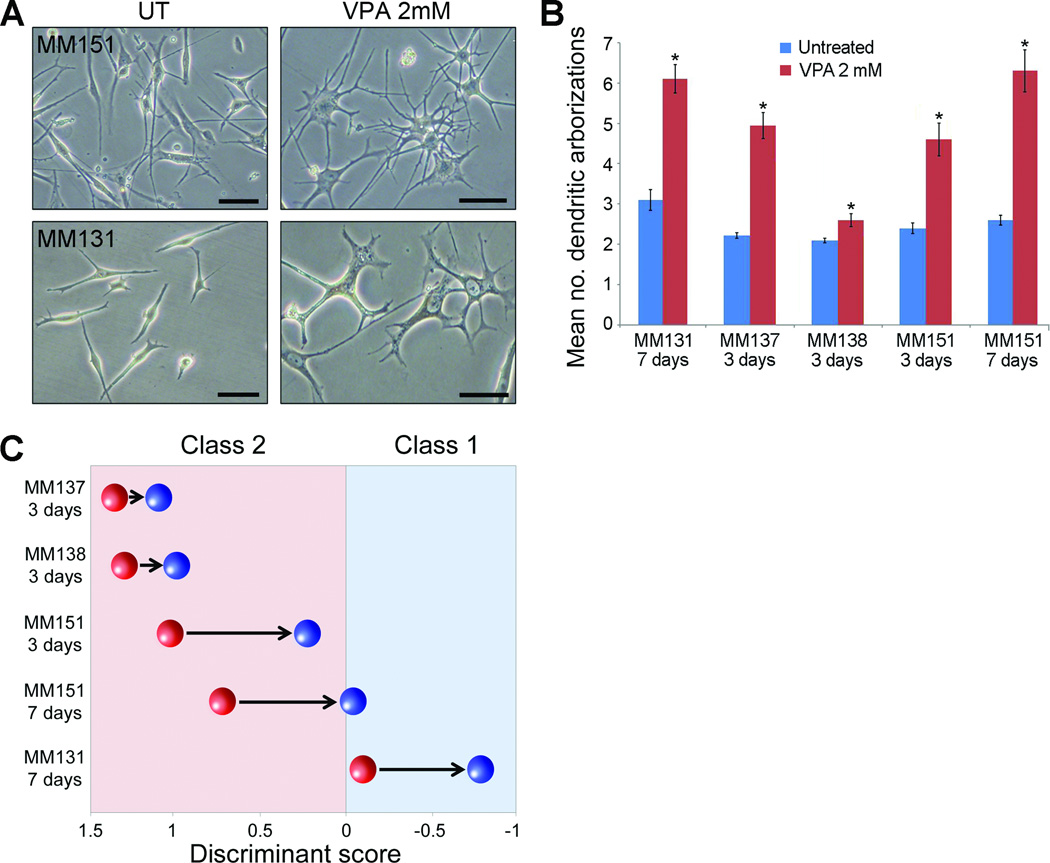
We then investigated whether HDAC inhibition could reverse the phenotypic consequences of BAP1 loss: loss of melanocytic differentiation and acquisition of the class 2 gene expression profile (6). Since UM cell lines undergo genetic and epigenetic artifacts in long term culture, for these experiments we used primary UM cells from five class 2 tumors (MM137, MM138, MM151, MM161 and MM162) and one class 1 tumor (MM131) which were obtained immediately after surgical resection and placed in short term culture. While only four of the class 2 samples contained detectable BAP1 mutations, all five showed low BAP1 RNA levels compared to the class 1 tumors (Supplementary Table S2 and Supplementary Fig. S3). In cells treated with VPA for 3 and 7 days, cell bodies became enlarged and flattened, and there was a marked increased in the number of dendritic arborizations, consistent with melanocytic differentiation (Fig. 5A–B). Untreated or VPA-treated cells were subjected to a multi-gene expression profile that classifies UMs as class 1 or class 2 (4). The results of this assay were reported as a SVM discriminant score; the more negative the score, the more class 1 the expression profile, the more positive the more class 2. VPA shifted all of the class 2 UM cells towards a class 1 profile, and it even shifted the class 1 UM cells towards a more class 1 profile (Fig. 5C and Supplementary Fig. S4). This effect was dose-dependent, and similar results were observed with LBH-589 (Supplementary Figs. S4 and S5C). The genes most affected by VPA were HTR2B (down-regulated by VPA) and LMCD1 (up-regulated by VPA) (Supplementary Fig. S5), both strong indicators of metastasizing UMs (4). A fourth HDAC inhibitor, SAHA, induced effects similar to VPA in UM cell lines and primary UM cells (Supplementary Fig. S6).
Figure 5. VPA induces differentiation and a shift to class 1 signature.
A, representative phase contrast images of primary UM cells from a class 2 tumor (MM151) and a class 1 tumor (MM131) untreated (UT) or VPA-treated for 7 days. Scale bar equal 50 µm. B, mean number of dendritic arborizations in primary cultured UM cells that were untreated (UT) or VPA-treated. C, Support vector machine (SVM) discriminant scores showing the effect of VPA treatment (2 mM) on gene expression signature. The more negative the number, the more class 1-like; the more positive the number, the more class 2-like. MM137, MM138 and MM151 cells were from class 2 tumors; MM131 was from a class 1 tumor. Red spheres represent the untreated cells and blue spheres the VPA-treated cells. *P<0.05.
DISCUSSION
Here we show that clinically achievable concentrations of several HDAC inhibitors can reprogram highly aggressive UM cells to a low grade, differentiated state. This is consistent with similar findings in other cancers (24). These findings suggest a potential role for HDAC inhibitors in patients with high risk class 2 UMs. Most of these patients harbor undetectable micrometastases at the time of primary eye tumor diagnosis, and the goal of HDAC inhibitor therapy would be to induce prolonged clinical dormancy (and longer patient survival) by driving these micrometastatic cells into a differentiated, quiescent state.
We have not yet found a UM cell line with naturally occurring BAP1 mutations (data not shown), despite many of these cell lines being derived from metastasizing UMs that likely contained cells with mutant BAP1. This suggests that BAP1-mutant UM cells may not grow well in culture. This has hindered our ability to study the effects of HDAC inhibitors on BAP1 mutant UM cells in vivo. However, in short term cultures, HDAC inhibitors did indeed reverse the biochemical, transcriptomic and morphologic consequences of BAP1 loss in primary UM cells. Knock down of BAP1 caused an increase in histone H2A ubiquitination, consistent with its H2A ubiquitin carboxy-terminal hydrolase activity (7, 22), which is critical for its tumor suppressor function (7, 8). HDAC inhibitors reversed the H2A hyperubiquitination caused by BAP1 loss, and they shifted the gene expression profile of class 2 cells towards a class 1 profile. Further, HDAC inhibitors induced morphologic changes consistent with melanocytic differentiation. These findings suggest that BAP1 normally functions at least in part to maintain melanocyte differentiation in UM cells, and that this function can be at least partially restored in the absence of BAP1 by increasing histone H3 acetylation. HDAC inhibitors reverse the biochemical defect caused by BAP1 loss (i.e., H2A hyperubiquitination), which may explain why BAP1-deficient UM cells were more sensitive to HDAC inhibition than BAP1-competent cells. However, since HDAC inhibitors act through multiple mechanisms of action, it is not surprising that they also had a differentiating effect on BAP1 wildtype tumor cells.
Histones undergo extensive post-translational modifications, including acetylation, methylation, phosphorylation, ubiquitination, and ribosylation, that comprise a complex combinatorial code that regulates gene expression (25). This system has been of particular interest in understanding the coordinated expression of transcriptional programs during development and differentiation. Among these modifications, acetylation has been one of the most thoroughly studied. The balance between acetylation and deacetylation is determined by the relative activities of histone acetyltransferases and HDACs, with increased acetylation promoting greater chromatin accessibility for gene expression. Histone ubiquitination is less well understood. Monoubiquitination of histone H2A on Lys119 plays an important role in X-chromosome inactivation, Polycomb group-dependent gene silencing and other developmental processes (26, 27). Distinct histone modifications also can function together in a coordinated fashion to regulate chromatin structure and gene expression. For example the histone deubiquitinase 2A-DUB regulates transcription by coordinating histone acetylation and deubiquitination, and destabilizing the association of linker histone H1 with nucleosomes (28). Further, inhibition of HDAC activity has been shown to decrease histone ubiquitination through transcriptional repression of the PRC1 component BMI1 (23). This may explain the efficacy of HDAC inhibitors in UM cells.
VPA and LBH-589 have both demonstrated therapeutic potential as monotherapy or in combination with other antitumor drugs in solid and hematologic malignancies (29). As expected, based on their mechanism of action, HDAC inhibitors have not been shown to be effective as a cytotoxic therapy in patients with advanced metastatic melanoma (30), but the findings herein suggest that they may have a role in preventing the progression of micrometastatic disease. We focused here on VPA because it had a more prominent effect on cell cycle arrest and differentiation, compared to TSA and LBH-589, which were more cytotoxic. The effect of SAHA was similar to VPA. VPA is a well characterized compound that has been used for almost 30 years in the treatment of epilepsy and more recently as an anti-cancer agent (29). The toxicity profile of VPA would allow its use as an adjuvant agent in high risk cancer patients. Taken together, these factors suggest that a clinical trial of adjuvant VPA or a similar HDAC inhibitor in high risk class 2 UM patients may be warranted. Gene expression profiling can allow enrollment of high risk class 2 patients, about half of whom will develop overt metastatic disease within 3 years of eye tumor diagnosis (4).
TRANSLATIONAL RELEVANCE.
Uveal melanoma is highly metastatic and, once it has disseminated, it is highly recalcitrant to available therapies. Recently, we discovered that inactivating mutations in the histone H2A ubiquitin hydrolase BAP1 are strongly associated with metastasis in uveal melanoma. In this study, we demonstrate that HDAC inhibitors reverse the biochemical effects of BAP1 loss, induce melanocytic differentiation and cell cycle arrest, and inhibit the growth of uveal melanoma tumors in vivo in a xenograft model. These findings suggest that HDAC inhibitors may be effective in an adjuvant setting for inducing differentiation and prolonging the dormancy of micrometastatic disease in uveal melanoma.
Supplementary Material
Supplementary Figure S1. HDAC inhibitors induce histone H3 hyperacetylation in UM cells. A, acetyl-histone H3 (Ac-H3) immunofluorescence of 92.1 and OCM1A cells untreated (UT) or HDAC inhibitor-treated for 72 hours. Magnification 200X. B, acetyl-histone H3 (Ac-H3, upper panel) and total histone H3 (lower panel) western blots of 92.1 cells untreated (UT) or HDAC inhibitor-treated.
Supplementary Figure S2. HDAC inhibitors induce differentiation of UM cells. HDAC inhibitors induce differentiation of UM cells. 92.1, OCM1A and Mel202 cell lines were either untreated (UT) or HDAC inhibitor-treated for 72 hours. A, representative phase contrast images of morphologic changes. Magnification 100X. B, spindle morphology index determined using the maximal cell length versus width ratio. *P<0.05.
Supplementary Figure S3. Cultured cells from class 2 tumors express low levels of BAP1 mRNA compared to those from class 1 tumors. RNA expression of BAP1 in primary UM cells derived from class 1 (MM131; blue dot) and class 2 (MM137, MM138, MM151, MM161, MM162; red dots) tumors measured by qPCR. The box-and-whiskers plots represent the expression of BAP1 in eight class 1 and twenty-eight class 2 primary UM tumors. Fold change was calculated using the sample with the lowest BAP1 expression as a calibrator. Middle line represents median; box, 25th to 75th percentiles; outer bars, minimum and maximum values.
Supplementary Figure S4. HDAC inhibitors shift class 2 UM cells and UM cell lines toward a class 1 expression signature in a dose-dependent fashion. A, Support vector machine (SVM) discriminant scores of primary UM cells treated with VPA (left panel) or LBH-589 (right panel) for 72 hours. B, SVM discriminant scores of UM cell lines treated with VPA for 72 hours. The more negative the number, the more class 1-like; the more positive the number, the more class 2-like. Black dots correspond to untreated cells and red dots to HDAC inhibitor-treated cells; x-axis represents HDAC inhibitor concentrations and y-axis represents discriminant score values in SVM units. *P<0.05, **P<0.005.
Supplementary Figure S5. Effects of HDAC inhibitors on gene expression signature. A, mRNA levels of the multi-gene prognostic assay of MM151 class 2 cells treated with increasing concentration of VPA for 72 hours (left panel) and of three class 2 cells (MM137, MM138, MM151) treated with 2 mM VPA for 72 hours (right panel), presented as relative change in class 2 treated/untreated cells. B, mRNA levels of the multi-gene clinical prognostic assay of MM151 class 2 cells (left panel) and MM131 class 1 cells (right panel) treated with 2 mM VPA for 7 days, presented as relative change in treated/untreated cells. C, mRNA levels of the multi-gene clinical prognostic assay of three class 2 cells (MM137, MM138, MM151) treated with LBH-589 for 72 hours, presented as relative change in class 2 treated/untreated cells. *P<0.05. Blue boxes indicate genes with higher expression in class 1 tumors, and red boxes indicate genes with higher expression in class 2 tumors.
Supplementary Figure S6. SAHA has similar effects to other HDAC inhibitors in UM cells. A, MTS cell viability assays after 72 hours treatment. The absorbance of control cells at 490 nm was taken as 100%. B, BrdU incorporation assays after 72 hours treatment. The absorbance at 370 nm of control cells was taken as 100%. C, cell cycle analysis by flow cytometry using propidium iodide staining. Cells were either untreated (UT) or SAHA-treated for 48 hours; x-axis represents DNA content and y-axis represents cell number. D, mean number of dendritic arborizations in primary cultured UM cells that were untreated (UT) or SAHA-treated for 72 hours. C, Support vector machine (SVM) discriminant scores showing the effect of SAHA treatment (1 µM) on gene expression signature. The more negative the number, the more class 1-like; the more positive the number, the more class 2-like. MM137, MM161 and MM162 cells were from class 2 tumors. Red spheres represent the untreated cells and blue spheres the SAHA-treated cells. *P<0.05.
ACKNOWLEDGEMENTS
Financial support: This work was supported by Fonds de la Recherche en Santé du Québec Postdoctoral Training Award (SL), NIH/NIDCDT32 Research Training Program for Otolaryngology (ZTK), Alvin J. Siteman Cancer Center Summer Undergraduate Research Fellowship program (RSL), NIH R01 AR050266 (AMB), NIH R01 CA125970 (JWH), NIH R01 EY13169 (JWH), Barnes-Jewish Hospital Foundation (JWH), Kling Family Foundation (JWH), Horncrest Foundation (JWH) and Research to Prevent Blindness (JWH), and by an unrestricted grant to the Department of Ophthalmology and Visual Sciences from a Research to Prevent Blindness, Inc. and the NIH Vision Core Grant P30 EY02687.
This work was not supported by Castle Biosciences, Inc.
The authors would like to thank William Eades, Jon Christopher Holley, and Jacqueline Hughes in the Siteman Cancer Center High Speed Sorter Core Facility (NCI Cancer Center Support Grant #P30 CA91842).
Footnotes
Disclosure of potential conflicts of interest: JWH and Washington University may receive income based on a license of related technology by the University to Castle Biosciences, Inc.
REFERENCES
- 1.Wikman H, Vessella R, Pantel K. Cancer micrometastasis and tumour dormancy. Apmis. 2008;116:754–770. doi: 10.1111/j.1600-0463.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- 2.Landreville S, Agapova OA, Harbour JW. Emerging insights into the molecular pathogenesis of uveal melanoma. Future Oncol. 2008;4:629–636. doi: 10.2217/14796694.4.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finger PT, Kurli M, Reddy S, Tena LB, Pavlick AC. Whole body PET/CT for initial staging of choroidal melanoma. Br J Ophthalmol. 2005;89:1270–1274. doi: 10.1136/bjo.2005.069823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–468. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18:191–200. doi: 10.1097/CMR.0b013e3283005270. [DOI] [PubMed] [Google Scholar]
- 6.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 8.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011 doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onken MD, Worley LA, Person E, Char DH, Bowcock AM, Harbour JW. Loss of heterozygosity of chromosome 3 detected with single nucleotide polymorphisms is superior to monosomy 3 for predicting metastasis in uveal melanoma. Clin Cancer Res. 2007;13:2923–2927. doi: 10.1158/1078-0432.CCR-06-2383. [DOI] [PubMed] [Google Scholar]
- 13.Landreville S, Agapova OA, Kneass ZT, Salesse C, Harbour JW. ABCB1 identifies a subpopulation of uveal melanoma cells with high metastatic propensity. Pigment Cell Melanoma Res. 2011;24:430–437. doi: 10.1111/j.1755-148X.2011.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 15.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 18.Delston RB, Matatall KA, Sun Y, Onken MD, Harbour JW. p38 phosphorylates Rb on Ser567 by a novel, cell cycle-independent mechanism that triggers Rb-Hdm2 interaction and apoptosis. Oncogene. 2011;30:588–599. doi: 10.1038/onc.2010.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 21.Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bommi PV, Dimri M, Sahasrabuddhe AA, Khandekar JD, Dimri GP. The polycomb group protein BMI1 is a transcriptional target of HDAC inhibitors. Cell Cycle. 2010;9 doi: 10.4161/cc.9.13.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. Embo J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Leeuwen F, van Steensel B. Histone modifications: from genome-wide maps to functional insights. Genome Biol. 2005;6:113. doi: 10.1186/gb-2005-6-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 28.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan J, Cang S, Ma Y, Petrillo RL, Liu D. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J Hematol Oncol. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocca A, Minucci S, Tosti G, Croci D, Contegno F, Ballarini M, et al. A phase I-II study of the histone deacetylase inhibitor valproic acid plus chemoimmunotherapy in patients with advanced melanoma. Br J Cancer. 2009;100:28–36. doi: 10.1038/sj.bjc.6604817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. HDAC inhibitors induce histone H3 hyperacetylation in UM cells. A, acetyl-histone H3 (Ac-H3) immunofluorescence of 92.1 and OCM1A cells untreated (UT) or HDAC inhibitor-treated for 72 hours. Magnification 200X. B, acetyl-histone H3 (Ac-H3, upper panel) and total histone H3 (lower panel) western blots of 92.1 cells untreated (UT) or HDAC inhibitor-treated.
Supplementary Figure S2. HDAC inhibitors induce differentiation of UM cells. HDAC inhibitors induce differentiation of UM cells. 92.1, OCM1A and Mel202 cell lines were either untreated (UT) or HDAC inhibitor-treated for 72 hours. A, representative phase contrast images of morphologic changes. Magnification 100X. B, spindle morphology index determined using the maximal cell length versus width ratio. *P<0.05.
Supplementary Figure S3. Cultured cells from class 2 tumors express low levels of BAP1 mRNA compared to those from class 1 tumors. RNA expression of BAP1 in primary UM cells derived from class 1 (MM131; blue dot) and class 2 (MM137, MM138, MM151, MM161, MM162; red dots) tumors measured by qPCR. The box-and-whiskers plots represent the expression of BAP1 in eight class 1 and twenty-eight class 2 primary UM tumors. Fold change was calculated using the sample with the lowest BAP1 expression as a calibrator. Middle line represents median; box, 25th to 75th percentiles; outer bars, minimum and maximum values.
Supplementary Figure S4. HDAC inhibitors shift class 2 UM cells and UM cell lines toward a class 1 expression signature in a dose-dependent fashion. A, Support vector machine (SVM) discriminant scores of primary UM cells treated with VPA (left panel) or LBH-589 (right panel) for 72 hours. B, SVM discriminant scores of UM cell lines treated with VPA for 72 hours. The more negative the number, the more class 1-like; the more positive the number, the more class 2-like. Black dots correspond to untreated cells and red dots to HDAC inhibitor-treated cells; x-axis represents HDAC inhibitor concentrations and y-axis represents discriminant score values in SVM units. *P<0.05, **P<0.005.
Supplementary Figure S5. Effects of HDAC inhibitors on gene expression signature. A, mRNA levels of the multi-gene prognostic assay of MM151 class 2 cells treated with increasing concentration of VPA for 72 hours (left panel) and of three class 2 cells (MM137, MM138, MM151) treated with 2 mM VPA for 72 hours (right panel), presented as relative change in class 2 treated/untreated cells. B, mRNA levels of the multi-gene clinical prognostic assay of MM151 class 2 cells (left panel) and MM131 class 1 cells (right panel) treated with 2 mM VPA for 7 days, presented as relative change in treated/untreated cells. C, mRNA levels of the multi-gene clinical prognostic assay of three class 2 cells (MM137, MM138, MM151) treated with LBH-589 for 72 hours, presented as relative change in class 2 treated/untreated cells. *P<0.05. Blue boxes indicate genes with higher expression in class 1 tumors, and red boxes indicate genes with higher expression in class 2 tumors.
Supplementary Figure S6. SAHA has similar effects to other HDAC inhibitors in UM cells. A, MTS cell viability assays after 72 hours treatment. The absorbance of control cells at 490 nm was taken as 100%. B, BrdU incorporation assays after 72 hours treatment. The absorbance at 370 nm of control cells was taken as 100%. C, cell cycle analysis by flow cytometry using propidium iodide staining. Cells were either untreated (UT) or SAHA-treated for 48 hours; x-axis represents DNA content and y-axis represents cell number. D, mean number of dendritic arborizations in primary cultured UM cells that were untreated (UT) or SAHA-treated for 72 hours. C, Support vector machine (SVM) discriminant scores showing the effect of SAHA treatment (1 µM) on gene expression signature. The more negative the number, the more class 1-like; the more positive the number, the more class 2-like. MM137, MM161 and MM162 cells were from class 2 tumors. Red spheres represent the untreated cells and blue spheres the SAHA-treated cells. *P<0.05.