Abstract
Activation of the KEAP1-NRF2 signaling pathway is an adaptive response to environmental and endogenous stresses and serves to render animals resistant to chemical carcinogenesis and other forms of toxicity, whereas disruption of the pathway exacerbates these outcomes. This pathway, which can be activated by sulfhydryl-reactive, small-molecule pharmacologic agents, regulates the inducible expression of an extended battery of cytoprotective genes, often by direct binding of the transcription factor to antioxidant response elements in the promoter regions of target genes. However, it is becoming evident that some of the protective effects may be mediated indirectly through cross talk with additional pathways affecting cell survival and other aspects of cell fate. These interactions provide a multi-tiered, integrated response to chemical stresses. This review highlights recent observations on the molecular interactions and their functional consequences between NRF2 and the arylhydrocarbon receptor (AhR), NF-κB, p53, and Notch1 signaling pathways. Antioxid. Redox Signal. 13, 1649–1663.
Introduction
NRF2 (NF-E2-related factor 2, NFE2L2) is a transcription factor that mediates a broad-based set of adaptive responses to intrinsic and extrinsic cellular stresses (73). As such, NRF2 influences sensitivity to physiologic and pathologic processes affected by oxidative and electrophilic stresses, such as those imposed by inflammation and exposures to environmental toxicants. Carcinogenesis, chronic obstructive pulmonary disease, obesogenesis, and neurodegeneration are known to be affected by the Nrf2 genotype in murine models (73).
Molecular details of the NRF2 signaling pathway have emerged over the past decade, as reviewed elsewhere in this issue. Notably, definition of the interacting partners of NRF2, such as KEAP1 (Kelch-like ECH-associated protein 1), and the means by which they sense and transduce chemical signals of stress are under intense investigation in many laboratories (48). Characterization of the immediate downstream target genes of NRF2 has also been well enumerated, although features that define the tissue and cell-type specificity in responses are poorly understood. The gene-expression responses to activation of NRF2 signaling, mediated through interactions of NRF2 and the antioxidant response element(s) (ARE: 5′-NTGAG/CNNNGC-3′) in the regulatory domains of its target genes, result in a distinctive cytoprotective response. Beyond the now-seminal response of catalyzing the detoxification of carcinogens and other xenobiotics through conjugation and trapping processes, genomic analyses indicated that gene families affected by Nrf2 (a) provide direct antioxidants; (b) encode enzymes that directly inactivate oxidants; (c) increase levels of glutathione synthesis and regeneration; (d) stimulate NADPH synthesis; (e) enhance toxin export through the multidrug-response transporters; (f ) enhance the recognition, repair, and removal of damaged proteins; (g) elevate nucleotide excision repair; (h) regulate expression of other transcription factors, growth factors and receptors, and molecular chaperones; and (i) inhibit cytokine-mediated inflammation (48, 73).
It also is clear, though, that these direct genomic responses do not account for all of the actions of pharmacologic agents that activate NRF2 signaling, nor do they explain the range of stress-response phenotypes observed when components of the pathway, notably, Nrf2 or Keap1, have been genetically disrupted in mice (162). Analyses of global transcriptional responses have hinted toward interactions between NRF2 signaling and other prominent signaling pathways. These interactions can occur in multiple forms. Post-translational modifications such as phosphorylation play a major role in the regulation of gene expression and function. These covalent modifications control intracellular distribution, transcriptional activity, and stability of transcription factors, including NRF2 (81, 143). Some transcription factors can antagonize NRF2 either by competing for binding to AREs or by inhibiting NRF2 through a physical association. Small MAF proteins, BACH1, and the immediate early proteins c-FOS and FRA1 can compete with NRF2 for binding to AREs (103, 151). Furthermore, several transcription factors, including activating transcription factor 3, proliferator-activated receptor (PPAR)γ, and retinoic acid receptor α have been reported to form inhibitory complexes with NRF2 (6, 13, 59, 155, 168). In this review, we focus on recent evidence for transcriptional cross-talk between NRF2 and the arylhydrocarbon receptor (AhR), NF-κB, p53, and Notch pathways, as summarized schematically in Fig. 1.
FIG. 1.
Possible means for regulation of cell survival and other cell-fate responses through interactions of NRF2 with additional cell-signaling pathways, including AhR, NF-κB, p53, and Notch1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
NRF2 Interactions with AhR Signaling
The aryl hydrocarbon receptor is a ligand-activated member of the bHLH/PAS (basic helix-loop-helix/Per-Arnt-Sim) family of transcription factors that mediates the biologic and toxic effects of its xenobiotic ligands (44). When bound by polycyclic aromatic hydrocarbons such as dioxins (TCDD), AhR translocates from the cytoplasm to the nucleus, heterodimerizes with AhR nuclear translocator (ARNT), and activates transcription through the xenobiotic-responsive element (XRE). The XRE consists of a canonic motif of 5′-TNGCGTG-3′. After nuclear export, AhR is degraded through the 26S proteasome pathway. It is well recognized that both AhR and NRF2 signaling regulate the expression of genes affecting the metabolism of xenobiotics. In some instances, the response elements recognizing these transcription factors can be found in the regulatory domains of the same target genes, such as Nqo1 (104). Ma et al. (95) reported that the inducible expression of NQO1 by the potent AhR ligand TCDD depends on both AhR and NRF2. Genetic experiments using Ahr-, Arnt-, or Nrf2-deficient cells revealed that induction of NQO1 by TCDD depended on the presence of AhR and ARNT, whereas the basal and inducible expression required a functional NRF2 pathway. Yaeger et al. (163) extended these findings by using Nrf2-deficient mice to demonstrate the requirement of NRF2 for the induction of Nqo1, Ugt1a6, and Gsta1 transcripts, among other genes, by TCDD in mouse liver. AhR–Nrf2 compound null mutant mice respond to neither AhR ligands nor prototypical NRF2 activators with target genes, such as Nqo1 (105). A functional collaboration between these pathways is also highlighted by the earlier findings of Talalay and colleagues (115), in which they described the roles of “monofunctional” and “bifunctional” activators of NRF2 signaling. “Monofunctional” inducers affect only NRF2 signaling. “Bifunctional inducers,” such as β-napthoflavone, may interact with the AhR directly to activate AhR-regulated genes, such as Cyp1a1, Cyp1a2, and Cyp1b1, and then undergo transformation by these enzymes to reactive intermediates that then trigger NRF2 signaling (see Fig. 2). In this way, a comprehensive set of responses to xenobiotic challenges can be mounted (79).
FIG. 2.
Regulation of xenobiotic metabolism by AhR and NRF2. Xenobiotics are metabolized by cytochrome P450s (CYPs) into intermediates that are reactive or nonreactive. Reactive intermediates can be metabolized by cytoprotective enzymes into less-toxic and often readily excretable products. When activated by a ligand, AhR in the cytoplasm complexes with ARNT and translocates into the nucleus and induces the transcription of CYPs by binding to the xenobiotic response elements (XREs) in the promoters of target genes. NRF2 bound to KEAP1 in the cytoplasm translocates into the nucleus when the pathway is activated by exogenous or endogenous electrophilic or free radical stresses, and binds to the antioxidant response elements (AREs) to induce transcription of cytoprotective genes.
The mechanisms underlying the linkages between AhR and NRF2 signaling need further investigation. Miao et al. (98) demonstrated that Nrf2 gene transcription is directly modulated by AhR activation. DNA sequence analyses of the murine Nrf2 promoter revealed an XRE located at −712 bp and two additional XRE-like motifs located further upstream. Direct binding of AhR to the Nrf2 promoter was observed. Moreover, silencing of AhR expression with siRNA obviated induction of Nrf2 mRNA by TCDD. NRF2 also autoregulates its own expression through an ARE-like element located in the proximal region of its promoter, leading to persistent nuclear accumulation of NRF2 and protracted induction of its target genes (83).
NRF2 also directly modulates AhR signaling, highlighting bidirectional interactions of these pathways (138). Constitutive expression of AhR was affected by Nrf2 genotype. Moreover, a pharmacologic activator of NRF2 signaling, CDDO-Im {1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole}, induced Ahr, Cyp1a1, and Cyp1b1 transcription in Nrf2+/+ mouse embryonic fibroblasts (MEFs) but not in Nrf2-/- MEFs. Reporter analysis and chromatin immunoprecipitation assays revealed that NRF2 directly binds to one ARE found in the −230 bp region of the promoter of Ahr. Better understanding of this bidirectional regulation of AhR and NRF2 pathways should provide clues on the roles of NRF2 in complex diseases and in uncharacterized phenotypes.
Cell-culture studies and knockout mouse models have shown that, in addition to controlling the xenobiotic detoxification response, AhR activation leads to G0/G1 arrest, diminished capacity for DNA replication, inhibition of cell proliferation, and impaired cell differentiation. AhR may function also as a tumor-suppressor gene that becomes silenced during the process of tumor formation (32). Transcriptional responses to ligand-dependent activation of AhR signaling vary across cell types and by species, but consistently influence cytochrome P450 and other monooxygenase activities (17). Ligand-dependent and -independent AhR signaling influences (a) positive regulation of transcription, vascular development, organ morphogenesis, and metabolic processes; (b) processes related to mRNA processing and transport; (c) processes related to cell death, proliferation, and regulation of apoptosis; (d) mitotic cell cycle; and (e) embryonic and other developmental processes (132).
Ahr-null mice and Nrf2-null mice share some common phenotypes. These transcription factors appear to function as cell-survival factors; both types of knockout mice exhibit increased sensitivity to infection (75, 147), carcinogenesis (32, 72, 117), altered responses to wounding (12, 18), and metabolic syndrome (137, 144). Studies on responses to adipogenic stimuli in vitro and high-fat stress diets in vivo have provided some opportunities for evaluating the physiological interactions between these pathways.
Shimba et al. (136) observed that TCDD treatment suppresses the conversion of 3T3-L1 cells into adipocytes. By using MEF cell lines derived from Ahr-knockout mice, Alexander et al. (5) reported that the AhR is a constitutive inhibitor of triglyceride synthesis and an early regulator of adipocyte differentiation. In addition, one of the phenotypes of Ahr-null mice is transient fatty liver (133), implying an in vivo regulatory role of AhR in the adipogenic process. Shin et al. (138) postulated that NRF2 would inhibit adipogenesis through the interaction with the AhR pathway. Nrf2-/- MEFs showed markedly accelerated adipogenesis on stimulation, whereas Keap1-/- MEFs (which exhibit higher NRF2 signaling) differentiated slowly compared with their congenic wild-type MEFs. Ectopic expression of AhR and dominant-positive Nrf2 in Nrf2-/- MEFs also substantially delayed differentiation. To evaluate how NRF2 regulates adipogenesis, differentiation markers involved at multiple stages of adipogenesis were analyzed. CEBPs (CCAAT-enhancer–binding proteins) and PPARs are the two families of transcription factors that play critical roles in adipogenesis (159). CEBPβ and CEBPδ function at an early phase of the differentiation process by sensing adipogenic stimuli and initiating expression of CEBPα and PPARγ (122). CEBPα and PPARγ play roles at a later stage by inducing and maintaining expression of adipocyte-specific genes, such as Fabp4. The elevation of CEBPβ on adipogenic stimulation is transient, whereas CEBPα and PPARγ remain upregulated for the duration of adipogenesis. Although the mechanism by which AhR regulates adipogenesis has not been fully characterized, recent work suggests that AhR affects differentiation stages that follow CEBPβ activation (i.e., CEBPα or PPARγ upregulation. In 3T3-L1 preadipocytes, forced expression of AhR resulted in lower induction of Fabp4 and Cebpα upon differentiation than did that in control cells, whereas the induction of Pparγ was not affected (136). Pparγ expression could be induced by differentiation in Ahr-/- MEFs, but levels were lower than those in Ahr+/+ MEFs (5). Similarly, mRNA levels of Cebpα and Fabp4 were higher in Nrf2-/- MEFs than in Nrf2+/+ MEFs, both before and after differentiation, whereas induction of Pparγ2 was not affected by the Nrf2 genotype. mRNA levels of Cebpβ were lower in Nrf2-/- MEFs than in Nrf2+/+ MEFs. In primary Keap1-/- MEFs, disruption of Keap1 resulted in minimal induction of Cebpα, Fabp4, and Pparγ2 upon differentiation. Cebpβ mRNA levels remained higher in Keap1-/- MEFs than in Keap1+/+ MEFs, both before and after induction of adipogenesis. Thus, NRF2 negatively modulates expression of Cebpα and Pparγ2 but not Cebpβ during the course of adipogenesis. The stages of differentiation that are affected by NRF2 directly overlap with those affected by AhR, thereby supporting the hypothesis that NRF2 inhibits adipogenesis through cross talk with AhR signaling (see Fig. 3).
FIG. 3.
Schematic representation of stages of adipogenesis regulated by AhR and NRF2. Adipogenic stimuli such as dexamethasone and isobutylmethylxanthine induce expression of the early markers CEBPβ and CEBPδ, which leads to induction of PPARγ and CEBPα. NRF2 induces expression of Ahr mRNA by directly binding to the promoter of the Ahr gene. AhR inhibits differentiation stages that follow CEBPβ activation (i.e., CEBPα or PPARγ upregulation).
Treatment with CDDO-Im effectively prevented high-fat diet–induced increases in body weight, adipose mass, and hepatic lipid accumulation in Nrf2 wild-type mice but not in Nrf2-disrupted mice (137). Wild-type mice on a high-fat diet and treated with CDDO-Im exhibited higher oxygen consumption and energy expenditure than did vehicle-treated mice, whereas food intake was lower in CDDO-Im–treated than in vehicle-treated mice. Levels of gene transcripts for fatty acid synthesis enzymes were downregulated after CDDO-Im treatment in the liver of wild-type mice. This inhibitory effect of CDDO-Im on lipogenic gene expression was significantly reduced in Nrf2-disrupted mice. However, the extent to which this pharmacologic activation of Nrf2 to produce an anti-obesogenic effect is mediated through AhR, if at all, is unclear.
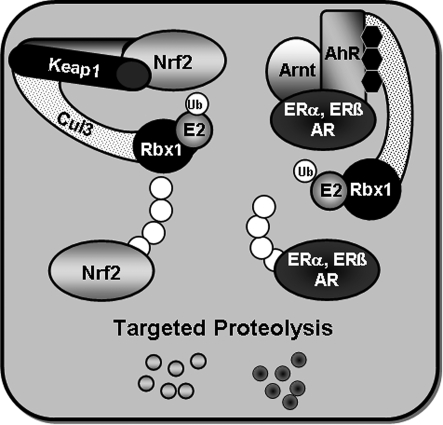
As with many transcription factors, the actions of the direct target genes of AhR alone do not fully explain its toxicologic and physiologic effects. In addition to interactions with NRF2 signaling, AhR extends its regulatory sweep by modulating the function of other transcription factors, including estrogen receptor (ERα and ERβ) and androgen receptor (AR). These cross-talk pathways are important mediators of the functions of endogenous and exogenous AhR ligands. The liganded AhR recently was shown to promote the ubiquitination and proteasomal degradation of ERs and AR by assembling a ubiquitin ligase complex, CUL4BAHR (108). Thus, complexes of the AhR with ERs or AR appear to regulate transcription as functional units by multiple mechanisms. As depicted in Fig. 4, KEAP1 also serves as an adaptor protein for an E3 ubiquitin ligase complex (78). Thus, broad similarities exist in the ways these pathways sense and respond to stresses. Indeed, other stress response–signaling pathways such as HIF-1α and NF-κB also interact with ubiquitin ligase systems to facilitate proteolytic turnover of signaling components (109).
FIG. 4.
E3 ubiquitin ligase activities of KEAP1 and AHR. KEAP1 binds with CUL3, RBX1, and E2 to facilitate the ubiquitination of NRF2, leading its enhanced degradation by the proteasome. Ligand-bound AhR assembles a CUL4B-based E3 ubiquitin ligase complex to mediate a nongenomic signaling pathway for influencing estrogen and androgen actions. ER, estrogen receptor; AR, androgen receptor.
Direct and Indirect NRF2–NF-κB Interactions
Cross talk between NRF2 and nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) is an area of extensive interest. NF-κB proteins are a family of transcription factors involved in several processes such as inflammation, immune response, apoptosis, development, and cell growth. Targets of NF-κB include genes classified as chemokines, cytokines, immunoreceptors, cell-adhesion molecules, stress-response genes, regulators of apoptosis, growth factors, and transcription factors, among many others. Tumor necrosis factor α (TNF-α), inducible NOS (iNOS), interleukin-1 (IL-1), intracellular adhesion molecule-1, and cyclooxygenase (COX-2) have all been shown to be induced by NF-κB (112). Pathologies related to alterations of the NF-κB pathway include allergies (25), Alzheimer disease (97), autoimmunity (46), obesity (40), atherosclerosis (125), arthritis (123), cancer (41), Crohn's disease (114), diabetes (164), and stroke (51).
All NF-κB proteins contain a conserved Rel homology domain responsible for dimerization and DNA binding to the consensus sequence GGGRNNYYCC, (R = purine, N = any base, Y = pyrimidine). The NF-κB family of proteins can be divided into two distinct groups based on the presence of a transactivation domain. RelA (p65), RelB, and c-Rel all contain transactivation domains, whereas p50 and p52 do not, and they require heterodimerization with the Rel proteins to activate transcription. Inactive NF-κB is located in the cytoplasm associated with the negative regulator IκB, which can conceal the nuclear-localization sequence or DNA-binding domain of NF-κB to prevent transcription (148, 157, 165). Activation of NF-κB is accomplished through phosphorylation of IκB by IκB kinases (IKKs), which leads to the release and nuclear translocation of NF-κB (22, 28, 150). A wide range of agents have the ability to stimulate IKK to activate NF-κB, including H2O2, TNF-α, IL-1, phorbol esters, hypoxia followed by reoxygenation, ultraviolet radiation, or microbial infection (112).
The NRF2 and NF-κB signaling pathways interface at several points to control the transcription or function of downstream target proteins. Many examples exist in which activation and repression occur between members of the two pathways through mechanisms of regulation ranging from direct effects on the transcription factors themselves to protein–protein interactions and second-messenger effects on target genes. Several cancer chemopreventive agents trigger NRF2 signaling with a concomitant repression of NF-κB and its target genes. As examples, sulforaphane [(-)-1-isothiocyanato-(4R)-methylsulfinyl)butane] reduces DNA binding of NF-κB and decreases generation of NO, PGE2, and TNF-α in Raw 264.7 macrophages without affecting IκB or nuclear translocation of the transcription factor (50). 3H-1,2-Dithiole-3-thione reduces the nuclear translocation and DNA binding of NF-κB, along with changes in phosphorylation of IκB in the hepatocytes of lipopolysaccharide (LPS)-treated rats (69). Epigallocatechin-3-gallate induces NRF2 and reduces NF-κB, TNF-α and IL-1β in the lungs of bleomycin-treated rats (140). Chalcone has been shown to induce NRF2 and inhibit the activation of NF-κB in endothelial cells (94). The synthetic triterpenoid CDDO-Me can inhibit NF-κB activity through repression of IKKβ (1). Curcumin is able to inhibit NF-κB by blocking phosphorylation and degradation of IκB in macrophages challenged with LPS (113).
NRF2 and NF-κB can interact to have direct effects on gene expression. NRF2 and NF-κB mediate the activation of IκB and ARE sites, respectively, through antagonism of transcription-factor binding to DNA. NF-κB was recently shown to prevent the transcription of NRF2-dependent genes by reducing available co-activator levels and promoting recruitment of a co-repressor. p65 and Nrf2 both bind to the CH1-KIX domain of CREB-binding protein (CBP), and after phosphorylation of p65 at Ser276, NF-κB is able to suppress transcription of ARE-dependent genes by preventing CBP from binding to Nrf2 (93). A second mechanism of p65 transcriptional repression of the ARE involving histone deacetylase 3 (HDAC3) has also been described. Overexpression of p65 causes the recruitment of HDAC3 to the ARE by binding to CBP or MafK. HDAC3 was shown to bind MafK in the NRF2 dimerization region and to prevent the acetylation of MafK by CBP (93). Further experiments showed that p65 is able to affect NRF2-dependent transcription through the recruitment of HDAC3 to the ARE by binding to CBP. HDAC3 was previously shown to bind and deacetylate CBP, resulting in reduced CBP-mediated transcriptional coactivation (24). A model for effects of reduced coactivator and recruitment of a co-repressor is depicted in Fig. 5. Phosphorylation of p65 on Ser276 reduces the available coactivator levels of CBP from NRF2, which causes decreased transcription through the ARE and also allows HDAC3 to bind CBP. The loss of CBP from the MafK-NRF2 heterodimer may then cause destabilization and allow HDAC3 to bind and deacetylate MafK. The HDAC could be then be recruited to the ARE through the action of the CBP-bound HDAC or a MafK homodimer–bound HDAC3 (93). Interestingly, this mechanism may be countered by induction of NRF2, which leads to increased cytoplasmic-to-nuclear translocation of the transcription factor. Experiments have shown that repression of NRF2-dependent transcription was eliminated by transfection of a dominant negative NRF2 that does not undergo cytoplasmic sequestration (93).
FIG. 5.
Repression of NRF2 signaling by members of the NF-κB pathway. p65 represses NRF2-dependent transcription at the ARE when phosphorylated on S276 by either competition for coactivator binding proteins or by recruiting HDAC to the ARE, which can deacetylate histone H4 and MafK.
Several experiments have shown increased NF-κB activation in Nrf2-/- mice when compared with wild-type after stimuli such as traumatic brain injury (65), LPS (147), TNF-α (147), ovalbumin (119), and respiratory syncytial virus (23). NRF2 has been implicated in NF-κB control through attenuation of phosphorylated IκB, which causes NF-κB degradation. Nrf2 -/- MEFs have higher levels of phosphorylated IκB and greater IKK kinase activity when compared with Nrf2 +/+ MEFs after challenge with LPS or TNF-α (147). Interestingly, overexpression of NRF2 seems to change only downstream NF-κB targets, not NF-κB or IκB (21, 68, 90). Overexpression of NRF2 in human aortic endothelial cells by using an adenoviral vector did not inhibit TNF-α–induced NF-κB activity or IκB degradation, but induction of monocyte chemoattractant protein-1 and VCAM-1 with TNF-α were inhibited (20). These inconsistencies could be due to the loss of NRF2, creating an environment with a diminished capacity to scavenge reactive oxygen species (ROS). The impaired expression of antioxidative genes could potentially lead to increased activation of the NF-κB pathway. Alternatively, enhancement of NF-κB due to direct transcriptional action of NRF2 may occur. In genetic studies, Nrf2-/- fibroblasts have shown reduced steady-state protein levels of p50 and p65 when compared with wild-type, whereas in vivo studies revealed that livers of Nrf2-/- mice exhibited greatly reduced transcript levels of p65 (161). By contrast, TNF-α or LPS stimulation of Nrf2-/- fibroblasts caused increased activation of NF-κB (147). Enhancement of NF-κB by NRF2 may be coordinated in a subset of the NF-κB family members to drive expression of specific genes. In Nrf2-/- fibroblasts p50 and p65 show reduced steady-state protein levels to 30% of wild type, whereas c-Rel protein levels were increased 400% (161). It is unknown whether NRF2 is able specifically to control expression of NF-κB proteins that lack a transactivation domain to repress transcription of NF-κB target proteins (88, 91). The enhancement of NRF2 due to direct transcriptional action of NF-κB may also be possible. In silico transcription factor–binding site analysis has identified an NF-κB consensus binding site in the murine promoter of NRF2 (102), which may increase transcription of NRF2 to blunt prolonged NF-κB activity. As multiple agents and stimuli such as ROS, LPS, flow shear stress, oxidized low-density lipoprotein, and cigarette smoke have been shown to induce both NRF2 and NF-κB activity (2, 7, 16, 42, 56, 77, 127), a completely antagonistic effect of the two transcription factors against one another in all situations is unlikely.
Interactions between downstream targets of NRF2 and NF-κB that ultimately lead to modulation of transcription factor activity have been discovered, which adds complexity to the two pathways. Three examples of NRF2 target genes that are able to influence NF-κB activity are heme oxygenase-1 (HO-1), NAD(P)H:quinone oxidoreductase (NQO1), and thioredoxin (TRX). HO-1, which breaks down heme to produce biliverdin, CO, and free iron, is able to modulate activation of NF-κB through the action of bilirubin and free iron (3, 66, 111, 135, 139, 145) (see Fig. 6). HO-1 has been shown specifically to prevent phosphorylation of RelA through the action of free iron at a site required for TNF-α–dependent NF-κB activation (135). Induction of HO-1 can inhibit IκB degradation, whereas inhibition of HO-1 increases p65 activity in HT-29 cells and colonic mucosa after treatment with TNF-α or IL-1ß (66). NQO1 has both positive and negative effects on NF-κB. NQO1 overexpression reduces LPS-induced expression of the downstream inflammatory genes TNF-α and IL-1 in THP-1 cells independent of NF-κB (128). Mice deficient of NQO1 in bone marrow, spleen, and thymus showed reduced NF-κB DNA binding under basal or LPS-treated conditions (61). TRX also has both positive and negative effects on NF-κB. TRX can modulate NF-κB activity through the reduction of cysteine residues (26, 36, 47, 74, 96, 116). Nuclear TRX is required for the reduction of cysteine residues on p50 to allow binding to DNA, whereas cytoplasmic TRX is able to block the degradation of IκB (54).
FIG. 6.
The interactions of the NRF2 and NF-κB target proteins HO-1 and iNOS and their products can regulate the activity of each other in macrophages to protect against an overabundance of NO. Excess NO acts as a signal to increase HO-1, which is able to scavenge NO and block the activity of iNOS to prevent further production.
Target proteins of the NRF2 and NF-κB pathways and their products can also interact to regulate enzyme activity. The NF-κB target IL-10 (15) has been shown to induce HO-1 in murine macrophages and in vivo (87). A feedback loop is present between HO-1 and iNOS in macrophages to protect the cell against injury due to excess NO. Expression of iNOS, which leads to overproduction of NO, causes induction of HO-1 (34, 60). HO-1 is then able to inhibit iNOS through the action of free iron (156) and CO (131), as well as through the overall reduction of heme (4) to prevent further production of NO (141). Interestingly, NO can be scavenged by biliverdin and bilirubin to further entangle the signaling pathways (71) (Fig. 6). HO-1 has also been shown negatively to affect VCAM-1 expression (9). Additional interactions between HO-1 and the NF-κB pathway can be seen with the HO-1 product CO. CO can repress the formation of TNF-α, IL-1β, and macrophage inflammatory protein-1β and increase IL-10 after LPS stimulation (110).
An NF-κB target that has been shown to influence NRF2 activity is COX-2 and its downstream product 15-deoxy Δ(12,14) prostaglandin J2 (15d-PGJ2). Induction of COX-2 by shear stress has been shown to inhibit phosphatidylinositol 3-kinase activity, causing reduction of the transcription of NRF2, NQO1, HO-1, GCLR, and GSTM1 in human chondrocytes. Restoration of NRF2-dependent induction of the genes was seen on treatment with the COX-2–specific inhibitors CAY10404 and NS398 (49). 15d-PGJ2 has been shown to play a role in NRF2 modulation by binding to KEAP1 and causing nuclear translocation of NRF2 in macrophages (63). Further evidence of a direct effect of 15d-PGJ2 on NRF2 activation was found in vivo by using Nrf2-/- mice and the COX-2–selective inhibitor NS-398 (99). 15d-PGJ2 has also been shown to provide negative feedback through its own pathway by binding IKK to prevent the activation of NF-κB in HeLa cells and appears to do the same in human peripheral blood monocytes (126).
NRF2 and NF-κB have a subset of target genes that are regulated through the actions of both transcription factors on ARE and κB sites. HO-1 has been shown to have a functional ARE that can be activated by NRF2 (3), as well as a functional NF-κB site, by using EMSA and DNase I footprinting analysis (86). Glutamate-cysteine ligase catalytic subunit (GCLC) can be activated by NRF2 through an ARE (101) or by NF-κB through a κB site after ethanol exposure (76). The inhibitory G protein, Gαi2, is another example of a gene activated by both NF-κB and NRF2 (8). Expression of the chemokine IL-8 can be increased by both NRF2 and NF-κB, but not through the usual mechanisms. NF-κB activates transcription of IL-8 by binding to the κB site (82), whereas NRF2 instead uses a mechanism that stabilizes IL-8 mRNA (166).
p53 and NRF2: Cooperation and Antagonism in Protection Against Cancer
The examination of p53 after its discovery in 1979 (92) has been extensive, with approximately 52,000 articles accessioned when “p53” is searched on PubMed. This investigative interest is not without reason, inasmuch as p53 is mutated in upward of 50% of human cancers through single, compound, or deletion mutations (55). This common frequency of mutations makes p53 an attractive target for chemotherapeutics. If p53 is to be targeted, however, the body of knowledge surrounding the protein should be expanded to provide full understanding of the interactions of p53 with other signaling molecules and networks.
p53 regulates a host of processes within the cell, both directly, including the induction of apoptosis through the permeabilization of the outer mitochondrial membrane through interaction with proapoptotic factors (43, 89, 100), and indirectly, through the transcription of genes that effect changes on the cell. Examples include both pro- and antiapoptotic responses, the induction of cellular senescence, and the repair of genotoxic damage (39). These various activities of p53 make it an integral part of the cellular defense against transformation: p53 controls cell fates by regulating the activation of apoptosis to remove a heavily damaged cell, induces terminal differentiation to remove the threat of a moderately damaged cell, or induces the transcription of cellular scavenging proteins to reestablish cellular homeostasis.
Iida et al. (58) showed that Nrf2 and p53 collaborate to protect against carcinogenesis (58), with the demonstrations that, compared with wild-type mice, either Nrf2 knockout mice or p53 heterozygotic mice are more susceptible to nitrosamine-induced bladder carcinogenesis, and that the Nrf2-/-::p53+/- compound knockout mice are even more susceptible. This interaction is not unexpected, as when either the cytoprotective response system of NRF2, or p53, a cell-fate decision molecule is removed, the incidence of cancer increases after carcinogen challenge (117, 146). Although broad explanations based on general understandings of pathways can be posited, the actual interactions between NRF2 and p53 in this setting and the collective effects on the expression and function of their downstream target genes remain to be established.
In a more-direct examination of the interaction between p53 and NRF2, p53 has been shown to influence NRF2-based transcription by Cimino and colleagues (33). Specifically, p53 suppresses the transcription of three NRF2 target genes driven through the ARE. These genes, x-ct, Nqo1, and Gst1α1, are all involved in mounting an antioxidant response within the cell, specifically neutralizing ROS. In the study, various cell lines were transfected with expression vectors containing Nrf2 or p53. In all circumstances, the transformation with both Nrf2 and p53 results in a reduction of transcript levels of the target genes when compared with cells transformed with Nrf2 alone. This, in combination with experiments in which cells were exposed to etoposide (a topoisomerase II–inhibiting chemotherapeutic drug) also shows that p53 has the ability to regulate negatively the NRF2 target gene transcripts. When p53 is highly upregulated, through either overexpression or a strong DNA-damaging agent, antioxidant response genes are downregulated through p53-mediated disruption. Whether or to what extent NRF2 regulates p53 signaling is not clear. Transcriptome analyses, along with unpublished observations (M.K. Kwak and N. Wakabayashi), suggest that NRF2 contributes to the basal expression of MDM2, an inhibitor of p53 (85). The potential additional level of p53 regulation through NRF2 allows the dampening of MDM2-based proteasomal degradation of p53 in circumstances in which p53 may be modulating the expression of NRF2 target genes. Dampening of Mdm2 would allow an enhanced p53 signal. This effect makes intuitive sense, as if p53 were mediating an apoptotic response through ROS production, NRF2-driven gene expression of anti-ROS enzymes would be counterproductive. This observation, however, is confounded by a recent article examining the relation between p21, a direct downstream target of p53, and NRF2 (19).
p21 is involved with many different cellular processes, including cell-cycle arrest, cell differentiation, senescence, apoptosis and DNA replication and repair, and oxidative stress (29, 31, 38, 86, 107). Chen et al. (19) recently showed a direct interaction between p21 and NRF2, which may explain the cytoprotective properties of p21 when faced with oxidative stress. These investigators demonstrated in vitro that p21 is able to interact with the DLG motif within NRF2, thereby attenuating KEAP1-based ubiquitination and subsequent proteasomal degradation. The antioxidative properties of p21 rely on the presence of NRF2, as an increase in p21 concentration leads to increased transcript levels of ARE-regulated genes, and the presence of p21 within cells yields an increase in the half-life of NRF2. Additionally, it was observed in vivo that both basal and induced levels of NRF2 protein are higher in p21 wild-type mice than in p21-null mice, with induced levels of NRF2 target gene proteins NQO1 and HO-1 being higher in p21 wild-type mice. This discovery contributes significantly to appreciation of the emerging network of signaling between p53 and NRF2 by demonstrating that a direct target of p53 upregulates NRF2 levels and activity.
The three puzzle pieces presented here come together in an intricate fashion, demonstrating a tunable response by p53 to induction by any number of stimuli, as shown in Fig. 7. When p53 is strongly induced, the induction of apoptosis through the expression of proapoptotic proteins through the generation of intracellular ROS is secured by the suppression of antioxidant gene expression regulated by Nrf2, as well as the suppression of basal levels of Mdm2, to ensure the strongest possible cell-death signal both through direct levels of p53 and by its direct activities remaining uncompromised. The upregulation of proapoptotic factors by p53 may rely on increased levels of intracellular ROS production for signaling. In this setting, ROS is not neutralized by NRF2 target genes such as Nqo1 and Gst1α1 because of transcriptional interference by p53 (33), and the proteasomal degradation of p53 is hampered because of the reduction in MDM2 levels.
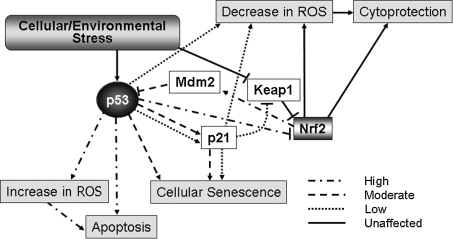
FIG. 7.
p53 modulates a gradient of responses to cellular stress. When the cell is exposed to a high level of stress, p53 acts in a proapoptotic fashion, modulating multiple pathways to secure a full commitment to cell death, whereas with a low level of cellular stress, p53 modifies gene expression to ensure cytoprotection after removal of the stress. The intermediate response is also possible, with p53 retaining the ability to induce cellular senescence through exit of the cell cycle and induction of terminal differentiation with a concurrent cytoprotective response.
On the other extreme, in the face of weak p53 induction, p21 is activated, stalling the cell cycle at the G1/S-phase checkpoint (30, 45, 160) This stalling allows the induction of DNA damage-repair functions by p53, the expression of proteins in an attempt to lower intracellular ROS (10, 129), and the stabilization of NRF2 by p21, thereby disrupting proteasomal degradation of NRF2 by KEAP1. This disruption of NRF2 turnover allows the nuclear translocation of NRF2, and the subsequent transcription of ARE-driven genes, leading to a general cytoprotective response. The collective actions of p53 and NRF2 lead to the repair of the cell and evasion of apoptosis. Links between NRF2 and the oxidative-stress–induced inhibition of proliferation have been well described (120, 121); in some instances, they may operate independent of p53.
The circumstance of a moderate induction of p53 allows some speculation that a middle ground exists between the promotion of programmed cell death and full cell repair and restoration of homeostasis, namely, that of cellular senescence through terminal differentiation. This balance is achieved through p21 activation after p53 induction through signaling. p21 blocks the cell cycle by stopping progression through the G1/S checkpoint (30, 45, 160), and allows the stabilization of NRF2 and an increased level of detoxification enzymes (19). p53 almost certainly plays a role in activating additional proteins that contribute to the process of rescuing a damaged cell that may be useful in a state of terminal differentiation but has experienced some insult that renders the cell a liability when used for mitotic purposes.
This system of dynamic response to cellular stresses by p53, p21, and NRF2 demonstrates a powerful mechanism for cellular and organismal maintenance, through the repair, differentiation, or destruction of damaged cells. In the case of mild damage, NRF2-mediated detoxification of carcinogens combined with a p21-mediated pause in the cell cycle and a p53/p21-mediated induction of DNA damage-repair enzymes salvages cells from damage, whereas in the case of extensive damage to a cell, p53 mediates the disruption of NRF2-mediated antioxidant response proteins, allowing p53-driven apoptosis, in toto reflecting a tunable response to environmental stresses.
NRF2 and NOTCH Signaling: Cell-Survival Factor Meets Cell-Fate Factor
Several lines of differential microarray analyses using RNAs isolated from MEFs prepared from Nrf2-/- and wild-type mice have shown that Notch1 and its direct downstream genes (Hes, Herp) (62) displayed decreased expression in Nrf2-/- MEF. Studies from our laboratory demonstrate that Notch1 signaling triggered through its activating ligands, DLL1 and JAG1, is dependent on Nrf2 genotype. Moreover, one or more functional ARE sequences exist in the promoter of Notch1. Therefore, it appears that NRF2 directly regulates Notch1 gene expression (153). It is now well established that signals through the NOTCH receptor are involved in the development of several cell types and that the modulation of these signals can markedly affect differentiation, proliferation, and apoptotic events. Genetic ablation studies indicate that Notch1 is crucial for early development and regrowth in a variety of tissues (57, 154). Activation of the pathway has been shown to be a potent inhibitor of differentiation in different developmental contexts and has been associated with the amplification of certain somatic stem cells (27, 35, 64, 80, 134, 142). Considering the significance of the NOTCH1 signal cascade in developmental biology, this indicated the possibility that NRF2 could be a key molecule affecting both embryonic and adult tissue stem cell renewal, as well as cell fate. Of course, this cross talk might not only be mediated by transcriptional machinery, as NOTCH signaling is regulated by various dynamic mechanisms, including posttranslational modifications (149). Critical to the evaluation of cross talk between transcription factors are assessments of their functional consequences. Several physiological lines of evidence support functional interactions between NRF2 and Notch1: liver regeneration, keratinogenesis, osteoblastogenesis, and adipogenesis (Fig. 8).
FIG. 8.
Possible effects of NRF2–Notch interactions on cell fate.
Nrf2-/- mice display an interesting phenotype in the process of liver regeneration (11). Nrf2-/- mice have been challenged with a two-thirds partial hepatectomy to view the impact of genotype on the kinetics of liver regeneration. These knockout mice showed a significant delay in the recovery of liver mass after partial hepatectomy. We have observed that this regeneration-defective phenotype is rescued by hepatocyte-specific expression of the NOTCH1 intracellular domain (NICD) in Nrf2-/- mice. NICD functions as a transcription co-factor of RBPJ. This finding provides clear evidence for the existence of NRF2-Notch1 cross talk within the damaged liver, and within the process of liver regeneration (153). In a rat model in which siRNAs against NOTCH1 and JAG1 (a NOTCH1 ligand) were injected into the superior mesenteric vein before the two-thirds partial hepatectomy, significantly suppressed proliferation of hepatocytes was reported at days 2 to 4 of the regenerative response (80). Apparently, the NRF2-NOTCH1 signaling pathway may be activated during liver regeneration and is potentially contributing to signals affecting cell growth and differentiation.
Keap1-/- mice, which exhibit hyperactivated NRF2 signaling, have a thickened cornified layer on the suprabasal layer of the esophagus (152), with a huge keratin mass derived from the cornified layer around the limiting ridge and cardia. All Keap1-null mice die within 3 weeks of malnutrition caused from esophageal and forestomach obstruction. Through microscopic observation, a clear difference is noted between the processes of cornified layer thickening in the esophagus versus the foregut. In the case of the esophagus, cell numbers in the basal layer seem to be unchanged, with the cornified layer exhibiting thickening. This appears to be a prodifferentiation mode of hyperkeratosis. Within the limiting ridge of the greater curvature and the cardiac portion of the forestomach, however, a proliferative form of hyperkeratosis is observed, with proliferation of undifferentiated cells. This phenotype might be explained by the excess Notch signaling produced by excessive NRF2 activation. NOTCH signaling is expressed in the gastrointestinal tract. Specifically, in both the esophagus and forestomach, NOTCH1 and JAG2 display the highest levels of expression within their families in the basal layer (70), where stem and progenitor cells are located (130). Progenitor cells are differentiated to keratinocytes in the suprabasal layer and denucleated in the cornified layer, with Notch signaling being weakly expressed in the suprabasal layer. Interestingly, NOTCH signaling induces terminal differentiation to activate p21 gene expression directly in keratinocytes (118, 158). This function of the NOTCH signal might be stronger in the esophagus and forestomach of Keap1-null mice.
Hinoi et al. (52) demonstrated that NRF2 negatively regulates osteoblast differentiation in the MC3T3-E1 cell system. They also showed that NRF2 might inhibit cellular differentiation and maturation in chondrocytes by using the pre-chondrogenic cell line ATDC5, derived from the mouse teratocarcinoma AT805 (53). As a proposed mechanism of these phenomena, they conjectured that the inhibition of RUNX2 (one of the essential factors for bone formation and chondral maturation) was caused by either direct or indirect interaction with NRF2. Conversely, Notch-signaling directly influences osteogenesis and chondrogenesis (14). Interestingly, these phenomena are all related to bHLH-transcription factors, especially HES and HERP, which are primary target genes of NOTCH signaling. These proteins function as negative regulators of target gene expression through direct cis-element (E-box, N-box)–binding machinery. They also facilitate proteasomal degradation through heterodimerization with other bHLH transcription factors. The heterodimer partners of HES or HERP primarily promote differentiation. Thus, they can lead to maintenance of current cell status. HES1 directly binds to the osteocalcin promoter and represses osteocalcin gene expression (167). Additionally, HES1/HERP2 physically interacts with RUNX2 in osteoblasts (37). It is also possible that NOTCH1 expression in osteoblasts is influenced by the NRF2 transcription factor, much as NOTCH1 is in MEF.
As discussed earlier, AhR and its downstream gene expression is reduced in Nrf2-/- MEF (138). This dampened gene expression impairs adipocyte differentiation from MEF. Nrf -/- MEF show markedly accelerated adipogenesis on stimulation, whereas Keap1-/- MEF, which demonstrate higher levels of NRF2 signaling, differentiate slowly compared with their congenic wild-type MEF. Within this system, ectopic expression of AhR rescues adipogenesis seen in Nrf2-/- MEF. However, recently Ross et al. (124) reported that HES1, a primary downstream gene product of NOTCH1 signaling, inhibits the differentiation of adipocytes from 3T3-L1 cells. They also demonstrated that both the bHLH and the WRPW (67) [which is a co-repressor (GROUCHO/TLE) binding motif ] domains in HES1 are required for its inhibitory effect. This study raised the possibility that the enhanced adipogenesis shown in Nrf2-/- MEF is caused by a weaker output of both NOTCH1 and AhR signaling than within wild-type MEF.
Conclusions
The NRF2 pathway has been studied in diverse contexts because of potential roles in stroke recovery, neurodegeneration, chronic obstructive pulmonary disease, and cancer prevention in multiple organs. The profiling of gene-expression changes mediated by the NRF2 pathway has been well documented. The interpretation of such studies, however, has been hampered by the inability to distinguish genes modulated directly by NRF2 from genes induced secondarily by NRF2-sensitive transcription factors and the mitigating impacts of tissue-specific responses. Nonetheless, it is clear that the protective effects of upregulation of NRF2 signaling can take several forms. Protection can be immediate, reflecting induction of genes directly regulated through NRF2 binding to AREs in target genes (e.g., the innate immune response and elevated cytoprotective responses to blunt cytokine surges or detoxify reactive intermediates, respectively) (73). The protective effects can be secondary through induction of macromolecular damage repair/removal systems (proteasome, DNA repair) (84, 106). Last, the protective effects can be tertiary through activation of tissue repair/regeneration pathways. In these latter cases, involvement in cross talk with additional pathways affecting cell survival and other aspects of cell fate most certainly play important collaborating roles. This review has highlighted several such collaborators (and sometime competitors): AhR, NF-κB, p53, and Notch1. Future studies evaluating the concordance between global mRNA expression profiles at the transcriptomic levels (i.e., oligonucleotide microarray analyses) and genome-wide NRF2-DNA binding analysis using high-throughput methods (i.e., chromatin-immunoprecipitation with parallel sequencing) should better establish a systems-level view of NRF2-dependent processes occurring within cells. Functional importance will require well-crafted molecular genetic studies in cell culture, and more important, in vivo, to define the critical cross-talk interactions between NRF2 and other transcription factors.
Abbreviations Used
- 15d-PGJ2
15-deoxy Δ(12,14) prostaglandin J2
- AhR
arylhydrocarbon receptor
- ARE
antioxidant response element
- ARNT
AhR nuclear translocator
- CBP
CREB-binding protein
- CDDO-Im
1-(2-cyano-3,12-dioxooleana-1,9[11]-dien-28-oyl)imidazole
- CEBPs
CCAAT-enhancer-binding proteins
- COX-2
cyclooxygenase-2
- GST
glutathione S-transferase
- HO-1
heme oxygenase-1
- IKK
IκB kinase
- IL-1
interleukin-1
- iNOS
inducible NOS
- KEAP1
Kelch-like ECH-associated protein 1
- LPS
lipopolysaccharide
- MEF
mouse embryonic fibroblast
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NICD
notch1 intracellular domain
- NQO1
NAD(P)H: quinone-acceptor 1
- NRF2
NF-E2–related factor 2
- PPAR
peroxisome proliferator-activated receptor
- ROS
reactive oxygen species
- Sulforaphane
(-)-1-isothiocyanato-(4R)-methylsulfinyl)butane
- TNF-α
tumor necrosis factor α
- TRX
thioredoxin
- XRE
xenobiotic-responsive element
Acknowledgments
This work was supported by NIH grants CA39416 and CA94076 (TWK) and the Maryland Cigarette Restitution Fund. SLS was supported by T32 ES07141 and T32 CA009110; JJS by T32 GM08763; and SS was the recipient of a Samsung Scholarship (Samsung Foundation of Culture).
References
- 1.Ahmad R. Raina D. Meyer C. Kharbanda S. Kufe D. Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on Cys-179. J Biol Chem. 2006;281:35764–35769. doi: 10.1074/jbc.M607160200. [DOI] [PubMed] [Google Scholar]
- 2.Ahn KS. Aggarwal BB. Transcription factor NF-kappaB: a sensor for smoke and stress signals. Ann N Y Acad Sci. 2005;1056:218–2133. doi: 10.1196/annals.1352.026. [DOI] [PubMed] [Google Scholar]
- 3.Alam J. Stewart D. Touchard C. Boinapally S. Choi AM. Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 4.Albakri QA. Stuehr DJ. Intracellular assembly of inducible NO synthase is limited by nitric oxide-mediated changes in heme insertion and availability. J Biol Chem. 1996;271:5414–5421. doi: 10.1074/jbc.271.10.5414. [DOI] [PubMed] [Google Scholar]
- 5.Alexander DL. Ganem LG. Fernandez-Salguero P. Gonzalez F. Jefcoate CR. Aryl-hydrocarbon receptor is an inhibitory regulator of lipid synthesis and of commitment to adipogenesis. J Cell Sci. 1998;111:3311–3322. doi: 10.1242/jcs.111.22.3311. [DOI] [PubMed] [Google Scholar]
- 6.Ansell PJ. Lo SC. Newton LG. Espinosa-Nicholas C. Zhang DD. Liu JH. Hannink M. Lubahn DB. Repression of cancer protective genes by 17beta-estradiol: ligand-dependent interaction between human Nrf2 and estrogen receptor alpha. Mol Cell Endocrinol. 2005;243:27–34. doi: 10.1016/j.mce.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Anwar AA. Li FY. Leake DS. Ishii T. Mann GE. Siow RC. Induction of heme oxygenase 1 by moderately oxidized low-density lipoproteins in human vascular smooth muscle cells: role of mitogen-activated protein kinases and Nrf2. Free Radic Biol Med. 2005;39:227–236. doi: 10.1016/j.freeradbiomed.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Arinze IJ. Kawai Y. Transcriptional activation of the human Galphai2 gene promoter through nuclear factor-kappaB and antioxidant response elements. J Biol Chem. 2005;280:9786–9795. doi: 10.1074/jbc.M414006200. [DOI] [PubMed] [Google Scholar]
- 9.Banning A. Brigelius-Flohe R. NF-kappaB, Nrf2, and HO-1 interplay in redox-regulated VCAM-1 expression. Antioxid Redox Signal. 2005;7:889–899. doi: 10.1089/ars.2005.7.889. [DOI] [PubMed] [Google Scholar]
- 10.Bensaad K. Tsuruta A. Selak MA. Vidal MN. Nakano K. Bartrons R. Gottlieb E. Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Beyer TA. Xu W. Teupser D. auf dem Keller U. Bugnon P. Hildt E. Thiery J. Kan YW. Werner S. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 2008;27:212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun S. Hanselmann C. Gassmann MG. auf dem Keller U. Born-Berclaz C. Chan K. Kan YW. Werner S. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol Cell Biol. 2002;22:5492–5505. doi: 10.1128/MCB.22.15.5492-5505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown SL. Sekhar KR. Rachakonda G. Sasi S. Freeman ML. Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2-related factor 2 (Nrf2)-regulated stress pathway. Cancer Res. 2008;68:364–368. doi: 10.1158/0008-5472.CAN-07-2170. [DOI] [PubMed] [Google Scholar]
- 14.Canalis E. Notch signaling in osteoblasts. Sci Signal. 2008;1:pe17. doi: 10.1126/stke.117pe17. [DOI] [PubMed] [Google Scholar]
- 15.Cao S. Zhang X. Edwards JP. Mosser DM. NF-kappaB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J Biol Chem. 2006;281:26041–26050. doi: 10.1074/jbc.M602222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carayol N. Chen J. Yang F. Jin T. Jin L. States D. Wang CY. A dominant function of IKK/NF-kappaB signaling in global lipopolysaccharide-induced gene expression. J Biol Chem. 2006;281:31142–31151. doi: 10.1074/jbc.M603417200. [DOI] [PubMed] [Google Scholar]
- 17.Carlson EA. McCulloch C. Koganti A. Goodwin SB. Sutter TR. Silkworth JB. Divergent transcriptomic responses to aryl hydrocarbon receptor agonists between rat and human primary hepatocytes. Toxicol Sci. 2009;112:257–272. doi: 10.1093/toxsci/kfp200. [DOI] [PubMed] [Google Scholar]
- 18.Carvajal-Gonzalez JM. Roman AC. Cerezo-Guisado MI. Rico-Leo EM. Martin-Partido G. Fernandez-Salguero PM. Loss of dioxin-receptor expression accelerates wound healing in vivo by a mechanism involving TGFbeta. J Cell Sci. 2009;122:1823–1833. doi: 10.1242/jcs.047274. [DOI] [PubMed] [Google Scholar]
- 19.Chen W. Sun Z. Wang XJ. Jiang T. Huang Z. Fang D. Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen XL. Dodd G. Thomas S. Zhang X. Wasserman MA. Rovin BH. Kunsch C. Activation of Nrf2/ARE pathway protects endothelial cells from oxidant injury and inhibits inflammatory gene expression. Am J Physiol Heart Circ Physiol. 2006;290:H1862–H1870. doi: 10.1152/ajpheart.00651.2005. [DOI] [PubMed] [Google Scholar]
- 21.Chen XL. Varner SE. Rao AS. Grey JY. Thomas S. Cook CK. Wasserman MA. Medford RM. Jaiswal AK. Kunsch C. Laminar flow induction of antioxidant response element-mediated genes in endothelial cells: a novel anti-inflammatory mechanism. J Biol Chem. 2003;278:703–711. doi: 10.1074/jbc.M203161200. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZJ. Parent L. Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 23.Cho HY. Imani F. Miller-DeGraff L. Walters D. Melendi GA. Yamamoto M. Polack FP. Kleeberger SR. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am J Respir Crit Care Med. 2009;179:138–150. doi: 10.1164/rccm.200804-535OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang HC. Chang CW. Chang GD. Yao TP. Chen H. Histone deacetylase 3 binds to and regulates the GCMa transcription factor. Nucleic Acids Res. 2006;34:1459–1469. doi: 10.1093/nar/gkl048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cousins DJ. McDonald J. Lee TH. Therapeutic approaches for control of transcription factors in allergic disease. J Allergy Clin Immunol. 2008;121:803–809. doi: 10.1016/j.jaci.2008.02.008. quiz 810–811. [DOI] [PubMed] [Google Scholar]
- 26.Das KC. c-Jun NH2-terminal kinase-mediated redox-dependent degradation of IkappaB: role of thioredoxin in NF-kappaB activation. J Biol Chem. 2001;276:4662–4670. doi: 10.1074/jbc.M006206200. [DOI] [PubMed] [Google Scholar]
- 27.de la Pompa JL. Wakeham A. Correia KM. Samper E. Brown S. Aguilera RJ. Nakano T. Honjo T. Mak TW. Rossant J. Conlon RA. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 28.DiDonato JA. Hayakawa M. Rothwarf DM. Zandi E. Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 29.Dotto GP. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta. 2000;1471:M43–M56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 30.el-Deiry WS. Tokino T. Velculescu VE. Levy DB. Parsons R. Trent JM. Lin D. Mercer WE. Kinzler KW. Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 31.Esposito F. Cuccovillo F. Russo L. Casella F. Russo T. Cimino F. A new p21waf1/cip1 isoform is an early event of cell response to oxidative stress. Cell Death Differ. 1998;5:940–945. doi: 10.1038/sj.cdd.4400427. [DOI] [PubMed] [Google Scholar]
- 32.Fan Y. Boivin GP. Knudsen ES. Nebert DW. Xia Y. Puga A. The aryl hydrocarbon receptor functions as a tumor suppressor of liver carcinogenesis. Cancer Res. 2010;70:212–222. doi: 10.1158/0008-5472.CAN-09-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faraonio R. Vergara P. Di Marzo D. Pierantoni MG. Napolitano M. Russo T. Cimino F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281:39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- 34.Foresti R. Clark JE. Green CJ. Motterlini R. Thiol compounds interact with nitric oxide in regulating heme oxygenase-1 induction in endothelial cells. Involvement of superoxide and peroxynitrite anions. J Biol Chem. 1997;272:18411–18417. doi: 10.1074/jbc.272.29.18411. [DOI] [PubMed] [Google Scholar]
- 35.Fre S. Huyghe M. Mourikis P. Robine S. Louvard D. Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 36.Freemerman AJ. Gallegos A. Powis G. Nuclear factor kappaB transactivation is increased but is not involved in the proliferative effects of thioredoxin overexpression in MCF-7 breast cancer cells. Cancer Res. 1999;59:4090–4094. [PubMed] [Google Scholar]
- 37.Garg V. Muth AN. Ransom JF. Schluterman MK. Barnes R. King IN. Grossfeld PD. Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 38.Gartel AL. Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- 39.Gatz SA. Wiesmuller L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 40.Gil A. Maria Aguilera C. Gil-Campos M. Canete R. Altered signalling and gene expression associated with the immune system and the inflammatory response in obesity. Br J Nutr. 2007;98(suppl 1):S121–S126. doi: 10.1017/S0007114507838050. [DOI] [PubMed] [Google Scholar]
- 41.Gilmore T. Gapuzan ME. Kalaitzidis D. Starczynowski D. Rel/NF-kappa B/I kappa B signal transduction in the generation and treatment of human cancer. Cancer Lett. 2002;181:1–9. doi: 10.1016/s0304-3835(01)00795-9. [DOI] [PubMed] [Google Scholar]
- 42.Go YM. Gipp JJ. Mulcahy RT. Jones DP. H2O2-dependent activation of GCLC-ARE4 reporter occurs by mitogen-activated protein kinase pathways without oxidation of cellular glutathione or thioredoxin-1. J Biol Chem. 2004;279:5837–5845. doi: 10.1074/jbc.M307547200. [DOI] [PubMed] [Google Scholar]
- 43.Green DR. Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu YZ. Hogenesch JB. Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 45.Harper JW. Adami GR. Wei N. Keyomarsi K. Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi T. Faustman D. Defective function of the proteasome in autoimmunity: involvement of impaired NF-kappaB activation. Diabetes Technol Ther. 2000;2:415–428. doi: 10.1089/15209150050194288. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi T. Ueno Y. Okamoto T. Oxidoreductive regulation of nuclear factor kappa B. Involvement of a cellular reducing catalyst thioredoxin. J Biol Chem. 1993;268:11380–11388. [PubMed] [Google Scholar]
- 48.Hayes JD. McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 49.Healy ZR. Lee NH. Gao X. Goldring MB. Talalay P. Kensler TW. Konstantopoulos K. Divergent responses of chondrocytes and endothelial cells to shear stress: cross-talk among COX-2, the phase 2 response, and apoptosis. Proc Natl Acad Sci U S A. 2005;102:14010–14015. doi: 10.1073/pnas.0506620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heiss E. Herhaus C. Klimo K. Bartsch H. Gerhauser C. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J Biol Chem. 2001;276:32008–32015. doi: 10.1074/jbc.M104794200. [DOI] [PubMed] [Google Scholar]
- 51.Herrmann O. Baumann B. de Lorenzi R. Muhammad S. Zhang W. Kleesiek J. Malfertheiner M. Kohrmann M. Potrovita I. Maegele I. Beyer C. Burke JR. Hasan MT. Bujard H. Wirth T. Pasparakis M. Schwaninger M. IKK mediates ischemia-induced neuronal death. Nat Med. 2005;11:1322–1329. doi: 10.1038/nm1323. [DOI] [PubMed] [Google Scholar]
- 52.Hinoi E. Fujimori S. Wang L. Hojo H. Uno K. Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J Biol Chem. 2006;281:18015–18024. doi: 10.1074/jbc.M600603200. [DOI] [PubMed] [Google Scholar]
- 53.Hinoi E. Takarada T. Fujimori S. Wang L. Iemata M. Uno K. Yoneda Y. Nuclear factor E2 p45-related factor 2 negatively regulates chondrogenesis. Bone. 2007;40:337–344. doi: 10.1016/j.bone.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 54.Hirota K. Murata M. Sachi Y. Nakamura H. Takeuchi J. Mori K. Yodoi J. Distinct roles of thioredoxin in the cytoplasm and in the nucleus: a two-step mechanism of redox regulation of transcription factor NF-kappaB. J Biol Chem. 1999;274:27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 55.Hollstein M. Sidransky D. Vogelstein B. Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 56.Hosoya T. Maruyama A. Kang MI. Kawatani Y. Shibata T. Uchida K. Warabi E. Noguchi N. Itoh K. Yamamoto M. Differential responses of the Nrf2-Keap1 system to laminar and oscillatory shear stresses in endothelial cells. J Biol Chem. 2005;280:27244–27250. doi: 10.1074/jbc.M502551200. [DOI] [PubMed] [Google Scholar]
- 57.Huppert SS. Le A. Schroeter EH. Mumm JS. Saxena MT. Milner LA. Kopan R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 58.Iida K. Itoh K. Maher JM. Kumagai Y. Oyasu R. Mori Y. Shimazui T. Akaza H. Yamamoto M. Nrf2 and p53 cooperatively protect against BBN-induced urinary bladder carcinogenesis. Carcinogenesis. 2007;28:2398–2403. doi: 10.1093/carcin/bgm146. [DOI] [PubMed] [Google Scholar]
- 59.Ikeda Y. Sugawara A. Taniyama Y. Uruno A. Igarashi K. Arima S. Ito S. Takeuchi K. Suppression of rat thromboxane synthase gene transcription by peroxisome proliferator-activated receptor gamma in macrophages via an interaction with NRF2. J Biol Chem. 2000;275:33142–33150. doi: 10.1074/jbc.M002319200. [DOI] [PubMed] [Google Scholar]
- 60.Immenschuh S. Tan M. Ramadori G. Nitric oxide mediates the lipopolysaccharide dependent regulation of the heme oxygenase-1 gene expression in cultured rat Kupffer cells. J Hepatol. 1999;30:61–69. doi: 10.1016/s0168-8278(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 61.Iskander K. Li J. Han S. Zheng B. Jaiswal AK. NQO1 and NQO2 regulation of humoral immunity and autoimmunity. J Biol Chem. 2006;281:30917–30924. doi: 10.1074/jbc.M605809200. [DOI] [PubMed] [Google Scholar]
- 62.Iso T. Kedes L. Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 63.Itoh K. Mochizuki M. Ishii Y. Ishii T. Shibata T. Kawamoto Y. Kelly V. Sekizawa K. Uchida K. Yamamoto M. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2) Mol Cell Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jensen CH. Jauho EI. Santoni-Rugiu E. Holmskov U. Teisner B. Tygstrup N. Bisgaard HC. Transit-amplifying ductular (oval) cells and their hepatocytic progeny are characterized by a novel and distinctive expression of delta-like protein/preadipocyte factor 1/fetal antigen 1. Am J Pathol. 2004;164:1347–1359. doi: 10.1016/S0002-9440(10)63221-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin W. Zhu L. Guan Q. Chen G. Wang QF. Yin HX. Hang CH. Shi JX. Wang HD. Influence of Nrf2 genotype on pulmonary NF-kappaB activity and inflammatory response after traumatic brain injury. Ann Clin Lab Sci. 2008;38:221–227. [PubMed] [Google Scholar]
- 66.Jun CD. Kim Y. Choi EY. Kim M. Park B. Youn B. Yu K. Choi KS. Yoon KH. Choi SC. Lee MS. Park KI. Choi M. Chung Y. Oh J. Gliotoxin reduces the severity of trinitrobenzene sulfonic acid-induced colitis in mice: evidence of the connection between heme oxygenase-1 and the nuclear factor-kappaB pathway in vitro and in vivo. Inflamm Bowel Dis. 2006;12:619–629. doi: 10.1097/01.ibd.0000225340.99108.8a. [DOI] [PubMed] [Google Scholar]
- 67.Kageyama R. Ohtsuka T. Tomita K. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol Cells. 2000;10:1–7. doi: 10.1007/s10059-000-0001-0. [DOI] [PubMed] [Google Scholar]
- 68.Kanninen K. Heikkinen R. Malm T. Rolova T. Kuhmonen S. Leinonen H. Yla-Herttuala S. Tanila H. Levonen AL. Koistinaho M. Koistinaho J. Intrahippocampal injection of a lentiviral vector expressing Nrf2 improves spatial learning in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:16505–10510. doi: 10.1073/pnas.0908397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karuri AR. Huang Y. Bodreddigari S. Sutter CH. Roebuck BD. Kensler TW. Sutter TR. 3H-1,2-dithiole-3-thione targets nuclear factor kappaB to block expression of inducible nitric-oxide synthase, prevents hypotension, and improves survival in endotoxemic rats. J Pharmacol Exp Ther. 2006;317:61–67. doi: 10.1124/jpet.105.096396. [DOI] [PubMed] [Google Scholar]
- 70.Katoh M. Katoh M. Notch signaling in gastrointestinal tract (review) Int J Oncol. 2007;30:247–251. [PubMed] [Google Scholar]
- 71.Kaur H. Hughes MN. Green CJ. Naughton P. Foresti R. Motterlini R. Interaction of bilirubin and biliverdin with reactive nitrogen species. FEBS Lett. 2003;543:113–119. doi: 10.1016/s0014-5793(03)00420-4. [DOI] [PubMed] [Google Scholar]
- 72.Kawajiri K. Kobayashi Y. Ohtake F. Ikuta T. Matsushima Y. Mimura J. Pettersson S. Pollenz RS. Sakaki T. Hirokawa T. Akiyama T. Kurosumi M. Poellinger L. Kato S. Fujii-Kuriyama Y. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci U S A. 2009;106:13481–13486. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kensler TW. Wakabayashi N. Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 74.Kim YC. Masutani H. Yamaguchi Y. Itoh K. Yamamoto M. Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf 2: a differential regulation of the antioxidant responsive element by a switch of its binding factors. J Biol Chem. 2001;276:18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 75.Kimura A. Naka T. Nakahama T. Chinen I. Masuda K. Nohara K. Fujii-Kuriyama Y. Kishimoto T. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med. 2009;206:2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimura T. Kawasaki Y. Okumura F. Sone T. Natsuki R. Isobe M. Ethanol-induced expression of glutamate-cysteine ligase catalytic subunit gene is mediated by NF-kappaB. Toxicol Lett. 2009;185:110–115. doi: 10.1016/j.toxlet.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 77.Knorr-Wittmann C. Hengstermann A. Gebel S. Alam J. Muller T. Characterization of Nrf2 activation and heme oxygenase-1 expression in NIH3T3 cells exposed to aqueous extracts of cigarette smoke. Free Radic Biol Med. 2005;39:1438–1448. doi: 10.1016/j.freeradbiomed.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 78.Kobayashi A. Kang MI. Okawa H. Ohtsuji M. Zenke Y. Chiba T. Igarashi K. Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kohle C. Bock KW. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73:1853–1862. doi: 10.1016/j.bcp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 80.Kohler C. Bell AW. Bowen WC. Monga SP. Fleig W. Michalopoulos GK. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 2004;39:1056–1065. doi: 10.1002/hep.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong AN. Owuor E. Yu R. Hebbar V. Chen C. Hu R. Mandlekar S. Induction of xenobiotic enzymes by the MAP kinase pathway and the antioxidant or electrophile response element (ARE/EpRE) Drug Metab Rev. 2001;33:255–271. doi: 10.1081/dmr-120000652. [DOI] [PubMed] [Google Scholar]
- 82.Kunsch C. Rosen CA. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol Cell Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kwak MK. Itoh K. Yamamoto M. Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kwak MK. Wakabayashi N. Greenlaw JL. Yamamoto M. Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kwak MK. Wakabayashi N. Itoh K. Motohashi H. Yamamoto M. Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway: identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 86.Lavrovsky Y. Schwartzman ML. Levere RD. Kappas A. Abraham NG. Identification of binding sites for transcription factors NF-kappa B and AP-2 in the promoter region of the human heme oxygenase 1 gene. Proc Natl Acad Sci U S A. 1994;91:5987–5991. doi: 10.1073/pnas.91.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee TS. Chau LY. Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat Med. 2002;8:240–246. doi: 10.1038/nm0302-240. [DOI] [PubMed] [Google Scholar]
- 88.Lernbecher T. Muller U. Wirth T. Distinct NF-kappa B/Rel transcription factors are responsible for tissue-specific and inducible gene activation. Nature. 1993;365:767–770. doi: 10.1038/365767a0. [DOI] [PubMed] [Google Scholar]
- 89.Letai AG. Diagnosing and exploiting cancer's addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- 90.Levonen AL. Inkala M. Heikura T. Jauhiainen S. Jyrkkanen HK. Kansanen E. Maatta K. Romppanen E. Turunen P. Rutanen J. Yla-Herttuala S. Nrf2 gene transfer induces antioxidant enzymes and suppresses smooth muscle cell growth in vitro and reduces oxidative stress in rabbit aorta in vivo. Arterioscler Thromb Vasc Biol. 2007;27:741–747. doi: 10.1161/01.ATV.0000258868.80079.4d. [DOI] [PubMed] [Google Scholar]
- 91.Lin R. Gewert D. Hiscott J. Differential transcriptional activation in vitro by NF-kappa B/Rel proteins. J Biol Chem. 1995;270:3123–3131. doi: 10.1074/jbc.270.7.3123. [DOI] [PubMed] [Google Scholar]
- 92.Linzer DI. Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 93.Liu GH. Qu J. Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta. 2008;1783:713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 94.Liu YC. Hsieh CW. Wu CC. Wung BS. Chalcone inhibits the activation of NF-kappaB and STAT3 in endothelial cells via endogenous electrophile. Life Sci. 2007;80:1420–1430. doi: 10.1016/j.lfs.2006.12.040. [DOI] [PubMed] [Google Scholar]
- 95.Ma Q. Kinneer K. Bi Y. Chan JY. Kan YW. Induction of murine NAD(P)H:quinone oxidoreductase by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires the CNC (cap 'n' collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem J. 2004;377:205–213. doi: 10.1042/BJ20031123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matthews JR. Wakasugi N. Virelizier JL. Yodoi J. Hay RT. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20:3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mattson MP. Camandola S. NF-kappaB in neuronal plasticity and neurodegenerative disorders. J Clin Invest. 2001;107:247–254. doi: 10.1172/JCI11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miao W. Hu L. Scrivens PJ. Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 99.Mochizuki M. Ishii Y. Itoh K. Iizuka T. Morishima Y. Kimura T. Kiwamoto T. Matsuno Y. Hegab AE. Nomura A. Sakamoto T. Uchida K. Yamamoto M. Sekizawa K. Role of 15-deoxy delta(12,14) prostaglandin J2 and Nrf2 pathways in protection against acute lung injury. Am J Respir Crit Care Med. 2005;171:1260–1266. doi: 10.1164/rccm.200406-755OC. [DOI] [PubMed] [Google Scholar]
- 100.Moll UM. Marchenko N. Zhang XK. p53 and Nur77/TR3: transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 101.Mulcahy RT. Wartman MA. Bailey HH. Gipp JJ. Constitutive and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J Biol Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 102.Nair S. Doh ST. Chan JY. Kong AN. Cai L. Regulatory potential for concerted modulation of Nrf2- and Nfkb1-mediated gene expression in inflammation and carcinogenesis. Br J Cancer. 2008;99:2070–2082. doi: 10.1038/sj.bjc.6604703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nguyen T. Huang HC. Pickett CB. Transcriptional regulation of the antioxidant response element: activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 104.Nioi P. Hayes JD. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat Res. 2004;555:149–171. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 105.Noda S. Harada N. Hida A. Fujii-Kuriyama Y. Motohashi H. Yamamoto M. Gene expression of detoxifying enzymes in AhR and Nrf2 compound null mutant mouse. Biochem Biophys Res Commun. 2003;303:105–111. doi: 10.1016/s0006-291x(03)00306-1. [DOI] [PubMed] [Google Scholar]
- 106.O'Dwyer PJ. Johnson SW. Khater C. Krueger A. Matsumoto Y. Hamilton TC. Yao KS. The chemopreventive agent oltipraz stimulates repair of damaged DNA. Cancer Res. 1997;57:1050–1053. [PubMed] [Google Scholar]
- 107.O'Reilly MA. Redox activation of p21Cip1/WAF1/Sdi1: a multifunctional regulator of cell survival and death. Antioxid Redox Signal. 2005;7:108–118. doi: 10.1089/ars.2005.7.108. [DOI] [PubMed] [Google Scholar]
- 108.Ohtake F. Baba A. Takada I. Okada M. Iwasaki K. Miki H. Takahashi S. Kouzmenko A. Nohara K. Chiba T. Fujii-Kuriyama Y. Kato S. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 109.Ohtake F. Fujii-Kuriyama Y. Kato S. AhR acts as an E3 ubiquitin ligase to modulate steroid receptor functions. Biochem Pharmacol. 2009;77:474–484. doi: 10.1016/j.bcp.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 110.Otterbein LE. Bach FH. Alam J. Soares M. Tao Lu H. Wysk M. Davis RJ. Flavell RA. Choi AM. Carbon monoxide has anti-inflammatory effects involving the mitogen-activated protein kinase pathway. Nat Med. 2000;6:422–428. doi: 10.1038/74680. [DOI] [PubMed] [Google Scholar]
- 111.Pae HO. Oh GS. Lee BS. Rim JS. Kim YM. Chung HT. 3-Hydroxyanthranilic acid, one of L-tryptophan metabolites, inhibits monocyte chemoattractant protein-1 secretion and vascular cell adhesion molecule-1 expression via heme oxygenase-1 induction in human umbilical vein endothelial cells. Atherosclerosis. 2006;187:274–284. doi: 10.1016/j.atherosclerosis.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 112.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 113.Pan MH. Lin-Shiau SY. Lin JK. Comparative studies on the suppression of nitric oxide synthase by curcumin and its hydrogenated metabolites through down-regulation of IkappaB kinase and NFkappaB activation in macrophages. Biochem Pharmacol. 2000;60:1665–1676. doi: 10.1016/s0006-2952(00)00489-5. [DOI] [PubMed] [Google Scholar]
- 114.Pena AS. Penate M. Genetic susceptibility and regulation of inflammation in Crohn's disease relationship with the innate immune system. Rev Esp Enferm Dig. 2002;94:351–360. [PubMed] [Google Scholar]
- 115.Prochaska HJ. Talalay P. Regulatory mechanisms of monofunctional and bifunctional anticarcinogenic enzyme inducers in murine liver. Cancer Res. 1988;48:4776–4782. [PubMed] [Google Scholar]
- 116.Qin J. Clore GM. Kennedy WM. Huth JR. Gronenborn AM. Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B. Structure. 1995;3:289–297. doi: 10.1016/s0969-2126(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 117.Ramos-Gomez M. Kwak MK. Dolan PM. Itoh K. Yamamoto M. Talalay P. Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rangarajan A. Talora C. Okuyama R. Nicolas M. Mammucari C. Oh H. Aster JC. Krishna S. Metzger D. Chambon P. Miele L. Aguet M. Radtke F. Dotto GP. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rangasamy T. Guo J. Mitzner WA. Roman J. Singh A. Fryer AD. Yamamoto M. Kensler TW. Tuder RM. Georas SN. Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reddy NM. Kleeberger SR. Bream JH. Fallon PG. Kensler TW. Yamamoto M. Reddy SP. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene. 2008;27:5821–5832. doi: 10.1038/onc.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reddy NM. Kleeberger SR. Yamamoto M. Kensler TW. Scollick C. Biswal S. Reddy SP. Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol Genomics. 2007;32:74–81. doi: 10.1152/physiolgenomics.00126.2007. [DOI] [PubMed] [Google Scholar]
- 122.Rosen ED. The transcriptional basis of adipocyte development. Prostaglandins Leukot Essent Fatty Acids. 2005;73:31–34. doi: 10.1016/j.plefa.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 123.Roshak AK. Callahan JF. Blake SM. Small-molecule inhibitors of NF-kappaB for the treatment of inflammatory joint disease. Curr Opin Pharmacol. 2002;2:316–321. doi: 10.1016/s1471-4892(02)00165-0. [DOI] [PubMed] [Google Scholar]
- 124.Ross DA. Hannenhalli S. Tobias JW. Cooch N. Shiekhattar R. Kadesch T. Functional analysis of Hes-1 in preadipocytes. Mol Endocrinol. 2006;20:698–705. doi: 10.1210/me.2005-0325. [DOI] [PubMed] [Google Scholar]
- 125.Ross JS. Stagliano NE. Donovan MJ. Breitbart RE. Ginsburg GS. Atherosclerosis: a cancer of the blood vessels? Am J Clin Pathol. 2001;1160(suppl):S97–S107. doi: 10.1309/YNCK-9R19-5JA3-K2K9. [DOI] [PubMed] [Google Scholar]
- 126.Rossi A. Kapahi P. Natoli G. Takahashi T. Chen Y. Karin M. Santoro MG. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IkappaB kinase. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 127.Rushworth SA. Chen XL. Mackman N. Ogborne RM. O'Connell MA. Lipopolysaccharide-induced heme oxygenase-1 expression in human monocytic cells is mediated via Nrf2 and protein kinase C. J Immunol. 2005;175:4408–4415. doi: 10.4049/jimmunol.175.7.4408. [DOI] [PubMed] [Google Scholar]
- 128.Rushworth SA. MacEwan DJ. O'Connell MA. Lipopolysaccharide-induced expression of NAD(P)H:quinone oxidoreductase 1 and heme oxygenase-1 protects against excessive inflammatory responses in human monocytes. J Immunol. 2008;181:6730–6737. doi: 10.4049/jimmunol.181.10.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sablina AA. Budanov AV. Ilyinskaya GV. Agapova LS. Kravchenko JE. Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sander GR. Powell BC. Expression of notch receptors and ligands in the adult gut. J Histochem Cytochem. 2004;52:509–516. doi: 10.1177/002215540405200409. [DOI] [PubMed] [Google Scholar]
- 131.Sarady JK. Zuckerbraun BS. Bilban M. Wagner O. Usheva A. Liu F. Ifedigbo E. Zamora R. Choi AM. Otterbein LE. Carbon monoxide protection against endotoxic shock involves reciprocal effects on iNOS in the lung and liver. FASEB J. 2004;18:854–856. doi: 10.1096/fj.03-0643fje. [DOI] [PubMed] [Google Scholar]
- 132.Sartor MA. Schnekenburger M. Marlowe JL. Reichard JF. Wang Y. Fan Y. Ma C. Karyala S. Halbleib D. Liu X. Medvedovic M. Puga A. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schmidt JV. Su GH. Reddy JK. Simon MC. Bradfield CA. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci U S A. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schroder N. Gossler A. Expression of Notch pathway components in fetal and adult mouse small intestine. Gene Expr Patterns. 2002;2:247–250. doi: 10.1016/s1567-133x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 135.Seldon MP. Silva G. Pejanovic N. Larsen R. Gregoire IP. Filipe J. Anrather J. Soares MP. Heme oxygenase-1 inhibits the expression of adhesion molecules associated with endothelial cell activation via inhibition of NF-kappaB RelA phosphorylation at serine 276. J Immunol. 2007;179:7840–7851. doi: 10.4049/jimmunol.179.11.7840. [DOI] [PubMed] [Google Scholar]
- 136.Shimba S. Wada T. Tezuka M. Arylhydrocarbon receptor (AhR) is involved in negative regulation of adipose differentiation in 3T3-L1 cells: AhR inhibits adipose differentiation independently of dioxin. J Cell Sci. 2001;114:2809–2817. doi: 10.1242/jcs.114.15.2809. [DOI] [PubMed] [Google Scholar]
- 137.Shin S. Wakabayashi J. Yates MS. Wakabayashi N. Dolan PM. Aja S. Liby KT. Sporn MB. Yamamoto M. Kensler TW. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shin S. Wakabayashi N. Misra V. Biswal S. Lee GH. Agoston ES. Yamamoto M. Kensler TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Soares MP. Seldon MP. Gregoire IP. Vassilevskaia T. Berberat PO. Yu J. Tsui TY. Bach FH. Heme oxygenase-1 modulates the expression of adhesion molecules associated with endothelial cell activation. J Immunol. 2004;172:3553–23563. doi: 10.4049/jimmunol.172.6.3553. [DOI] [PubMed] [Google Scholar]
- 140.Sriram N. Kalayarasan S. Sudhandiran G. Epigallocatechin-3-gallate augments antioxidant activities and inhibits inflammation during bleomycin-induced experimental pulmonary fibrosis through Nrf2-Keap1 signaling. Pulm Pharmacol Ther. 2009;22:221–236. doi: 10.1016/j.pupt.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 141.Srisook K. Cha YN. Super-induction of HO-1 in macrophages stimulated with lipopolysaccharide by prior depletion of glutathione decreases iNOS expression and NO production. Nitric Oxide. 2005;12:70–79. doi: 10.1016/j.niox.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 142.Stier S. Cheng T. Dombkowski D. Carlesso N. Scadden DT. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99:2369–2378. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]
- 143.Surh YJ. Kundu JK. Na HK. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008;74:1526–1539. doi: 10.1055/s-0028-1088302. [DOI] [PubMed] [Google Scholar]
- 144.Tanaka Y. Aleksunes LM. Yeager RL. Gyamfi MA. Esterly N. Guo GL. Klaassen CD. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther. 2008;325:655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- 145.Tenhunen R. Marver HS. Schmid R. The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proc Natl Acad Sci USA. 1968;61:748–755. doi: 10.1073/pnas.61.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tennant RW. French JE. Spalding JW. Identifying chemical carcinogens and assessing potential risk in short-term bioassays using transgenic mouse models. Environ Health Perspect. 1995;103:942–950. doi: 10.1289/ehp.95103942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Thimmulappa RK. Lee H. Rangasamy T. Reddy SP. Yamamoto M. Kensler TW. Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thompson JE. Phillips RJ. Erdjument-Bromage H. Tempst P. Ghosh S. I kappa B-beta regulates the persistent response in a biphasic activation of NF-kappa B. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 149.Tien AC. Rajan A. Bellen HJ. A Notch updated. J Cell Biol. 2009;184:621–629. doi: 10.1083/jcb.200811141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Traenckner EB. Pahl HL. Henkel T. Schmidt KN. Wilk S. Baeuerle PA. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Venugopal R. Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wakabayashi N. Itoh K. Wakabayashi J. Motohashi H. Noda S. Takahashi S. Imakado S. Kotsuji T. Otsuka F. Roop DR. Harada T. Engel JD. Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 153.Wakabayashi N. Shin S. Slocum S. Agoston ES. Wakabayashi J. Kwak MK. Misra V. Biswal S. Yamamoto M. Kensler TW. Regulation of Notch1 signaling by Nrf2: implications for tissue regeneration. Sci Signal. doi: 10.1126/scisignal.2000762. (in press, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang XD. Shou J. Wong P. French DM. Gao WQ. Notch1-expressing cells are indispensable for prostatic branching morphogenesis during development and re-growth following castration and androgen replacement. J Biol Chem. 2004;279:24733–24744. doi: 10.1074/jbc.M401602200. [DOI] [PubMed] [Google Scholar]
- 155.Wang XJ. Hayes JD. Henderson CJ. Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci USA. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Weiss G. Werner-Felmayer G. Werner ER. Grunewald K. Wachter H. Hentze MW. Iron regulates nitric oxide synthase activity by controlling nuclear transcription. J Exp Med. 1994;180:969–976. doi: 10.1084/jem.180.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Whiteside ST. Epinat JC. Rice NR. Israel A. I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wilson A. Radtke F. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 2006;580:2860–2868. doi: 10.1016/j.febslet.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 159.Wu Z. Bucher NL. Farmer SR. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol Cell Biol. 1996;16:4128–4136. doi: 10.1128/mcb.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xiong Y. Hannon GJ. Zhang H. Casso D. Kobayashi R. Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 161.Yang H. Magilnick N. Lee C. Kalmaz D. Ou X. Chan JY. Lu SC. Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kappaB and AP-1. Mol Cell Biol. 2005;25:5933–5946. doi: 10.1128/MCB.25.14.5933-5946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yates MS. Tran QT. Dolan PM. Osburn WO. Shin S. McCulloch CC. Silkworth JB. Taguchi K. Yamamoto M. Williams CR. Liby KT. Sporn MB. Sutter TR. Kensler TW. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–1031. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yeager RL. Reisman SA. Aleksunes LM. Klaassen CD. Introducing the “TCDD-inducible AhR-Nrf2 gene battery.”. Toxicol Sci. 2009;111:2382–2346. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yuan M. Konstantopoulos N. Lee J. Hansen L. Li ZW. Karin M. Shoelson SE. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of IKKbeta. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 165.Zabel U. Baeuerle PA. Purified human I kappa B can rapidly dissociate the complex of the NF-kappa B transcription factor with its cognate DNA. Cell. 1990;61:255–265. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]
- 166.Zhang X. Chen X. Song H. Chen HZ. Rovin BH. Activation of the Nrf2/antioxidant response pathway increases IL-8 expression. Eur J Immunol. 2005;35:3258–3267. doi: 10.1002/eji.200526116. [DOI] [PubMed] [Google Scholar]
- 167.Zhang Y. Lian JB. Stein JL. van Wijnen AJ. Stein GS. The Notch-responsive transcription factor Hes-1 attenuates osteocalcin promoter activity in osteoblastic cells. J Cell Biochem. 2009;108:651–659. doi: 10.1002/jcb.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou W. Lo SC. Liu JH. Hannink M. Lubahn DB. ERRbeta: a potent inhibitor of Nrf2 transcriptional activity. Mol Cell Endocrinol. 2007;278:52–62. doi: 10.1016/j.mce.2007.08.011. [DOI] [PubMed] [Google Scholar]