Abstract
Vertebrate genomes each encode hundreds of micro-RNAs (miRNAs), yet for few of these miRNAs is there empirical evidence as to which mRNA(s) they regulate. Here we report the identification of human lin-28 mRNA as a regulatory target of human miR-125b and its homolog miR-125a. Studies of miR-125b function in mouse P19 embryonal carcinoma cells induced to develop into neurons suggest a role for this regulatory miRNA in mammalian neuronal differentiation, since its increased concentration in these cells contributes to lin-28 downregulation. Within the lin-28 3′ untranslated region (UTR) are two conserved miRNA responsive elements (miREs) that mediate repression by miR-125b and miR-125a. Simultaneous deletion of both miREs renders the lin-28 3′ UTR almost completely insensitive to these miRNAs, indicating that these two miREs are the principal elements in the lin-28 3′ UTR that respond to miR-125. At the 3′ end of each element is an adenosine residue that makes a significant contribution to function irrespective of its complementarity to the 5′-terminal nucleotide of miR-125. By contrast to most earlier reports of gene repression by other miRNAs that are imperfectly complementary to their targets, lin-28 downregulation by miR-125 involves reductions in both translational efficiency and mRNA abundance. The decrease in the mRNA concentration is achieved by a posttranscriptional mechanism that is independent of the inhibitory effect on translation.
The cells of diverse eukaryotic organisms from animals to plants contain many different micro-RNAs (miRNAs), single-stranded oligoribonucleotides ∼22 nucleotides (nt) in length that function in gene regulation (22, 24, 45). In animal cells, miRNAs repress translation by annealing to mRNAs to which they are imperfectly complementary. Only a few mRNAs that are regulated by miRNAs in animals have been identified definitively, and little is known about the molecular mechanism by which they are repressed.
In animals, miRNAs are generated by processing of long primary transcripts (pri-miRNAs) by two RNase III-like endonucleases, Drosha and Dicer (8, 12, 18, 25). pri-miRNA cleavage by Drosha releases a 60- to 70-nt stem-loop intermediate (pre-miRNA) that is subsequently cut by Dicer to generate an RNA duplex comprising the miRNA and a complementary strand. The miRNA is then assembled as a single strand into a ribonucleoprotein complex and delivered to its mRNA targets. These miRNPs appear to be related, at least in part, to the ribonucleolytic complexes responsible for RNA interference, which contain ∼22-nt silencing RNAs (siRNAs) that guide mRNA cleavage at sites to which they are perfectly complementary (7, 10, 33, 37, 55). The similarities between translational repression by partially complementary miRNAs and nucleolytic silencing by fully complementary siRNAs have led to the proposal that they may represent divergent branches of a unified pathway for genetic control (11).
Gene regulation by naturally occurring miRNAs was first observed in Caenorhabditis elegans, where the miRNA products of the lin-4 and let-7 genes were shown to repress heterochronic genes involved in development (9, 24, 35, 44, 52). For example, lin-4 controls the expression of the lin-14, lin-28, and hbl-1 genes of C. elegans, targeting the mRNA transcripts of these genes for downregulation by pairing with imperfectly complementary sites in the 3′ untranslated region (UTR) and inhibiting translation (31, 40, 47, 52). By repressing these genes at specific times during larval development, lin-4 ensures the proper timing of cell fate transitions important for morphogenesis. In the case of the C. elegans lin-28 gene, which encodes a putative RNA-binding protein, expression is high in the first larval stage and then declines markedly in response to lin-4 and a second regulatory circuit that is lin-4 independent (35, 47). Gain-of-function mutations in lin-28 that abolish lin-4 repression cause a retarded developmental phenotype in various cell lineages, whereas loss-of-function mutations result in precocious development (1, 35).
Much less is known about the function of miRNAs in higher organisms, such as vertebrates, where the low degree of complementarity between miRNAs and their mRNA targets and an incomplete understanding of the rules governing miRNA function have complicated the identification of genes regulated in this manner. Indeed, despite the presence of hundreds of different miRNAs in vertebrate cells (2, 22, 29, 37, 42) and sequence-based predictions of possible regulatory targets (14, 19, 27, 28, 50), few such targets of vertebrate miRNA regulation have yet been verified empirically (15, 20, 28, 38, 39, 43, 54, 57), and for fewer still has a natural change in miRNA abundance been shown to modulate gene expression by directly targeting a specific mRNA in response to a regulatory signal.
Here we report the identification of the mammalian lin-28 gene as a regulatory target of miR-125b, a mammalian micro-RNA homologous to lin-4. Within the 3′ UTR of human and mouse lin-28 are two conserved miRNA-responsive elements (miREs) that are principally responsible for mediating the repressive effect of miR-125b and its paralog miR-125a. In mouse P19 embryonal carcinoma cells induced to differentiate into neurons, a marked increase in miR-125b abundance helps to downregulate lin-28 gene expression. Interestingly, the mechanism by which miR-125b represses lin-28 in mammalian cells appears to involve reductions not only in translational efficiency but also in mRNA abundance.
MATERIALS AND METHODS
DNA and RNA used for transfection.
Reporter plasmid pCMV-Luc, which encodes an mRNA comprising a firefly luciferase translational unit fused to the simian virus 40 (SV40) late 3′ UTR, was constructed from pGL3-control (Promega) by replacing the SV40 promoter with a cytomegalovirus (CMV) immediate-early promoter. In plasmid pCL-L28, the SV40 3′ UTR of pCMV-Luc was replaced with the 3′ UTR of human lin-28. Plasmids pCL-L28Δ1, pCL-L28Δ2, pCL-L28Δ0, pCL-L28Δ1+2, pCL-L28Δ0+1+2, pCL-L28ΔL7. and pCL-L28Δ0+1+2+L7 were derived from plasmid pCL-L28 by deleting a 15-bp segment encoding miRE1 (CACTGTGTTCTCAGG), an 18-bp segment encoding miRE2 (CATGAGCAATCTCAGGGA), a 13-bp segment encoding miRE0 (CATGTATCTCAGG), and/or a 27-bp segment encoding L7 (GAGTGCACAGCCTATTGAACTACCTCA). In other derivatives of pCL-L28, lin-28 3′ UTR segments with imperfect complementarity to miR-9 (TTTACTGCTAAAAACCAAAG), miR-30 (TCCGTGTTCTTTGGGGGTTTTGTTTACA), or miR-128 (GAGATCACCGCAAACCTACCTTACTGTG) were deleted. Plasmid pCL was constructed from pCMV-Luc by inserting a 0.11-kb spacer derived from the human immunodeficiency virus type 1 env gene between the luciferase stop codon and an XbaI site located just downstream of it. Plasmids pCL-2E1, pCL-4E1, and pCL-6E1 were constructed by inserting two, four, or six tandem copies of human lin-28 miRE1 (TCCTGCACTGTGTTCTCAGGTACAT) into the XbaI site of pCL. Plasmids pCL-2E2, pCL-4E2, pCL-6E2, pCL-2E0, pCL-4X, pCL-6X, and pCL-2Y were constructed similarly, except that multiple copies of miRE2 (AGGTACATGAGCAATCTCAGGGATAGCC), miRE0 (ACTGCCATGTATCTCAGGCTTGG), element X (CCAAATGCAAGTGAGGGTTCTGGGGGCAACC), or element Y (TTCTGTGGAAGGAGATCTCTCAGGAGTAA) were inserted into pCL. Plasmids pCL-BGH-con, pCL-BGHmut-con, pCL-BGH-6E1, and pCL-BGHmut-6E1 were constructed by inserting a 0.23-kb pcDNA3 gene fragment (Invitrogen) containing a wild-type (AATAAA) or mutated (TTCTTT) bovine growth hormone polyadenylation signal into pCL-con or pCL-6E1 at an EcoRI site located 10 bp downstream of the luciferase coding region. Plasmid pCL-6E1+hp was derived from pCL-6E1 by inserting the sequence CGGGGCGCGTGGTGGCGGCTGCAGCCGCCACCACGCGCCCCGGACGCGTAGCT at a HindIII site 30 bp upstream of the luciferase initiation codon. Plasmid pBC21/CMV/β-Gal (56), which encodes β-galactosidase, was used as an internal standard.
Plasmid pMIR125a, which was used to transfect 293T cells, was constructed from pCMV-Luc by replacing the luciferase translational unit between the CMV promoter and the SV40 3′ UTR with a 1.13-kb fragment of human chromosome 19 that encodes miR-125a flanked upstream by 166 nt and downstream by 941 nt. In plasmid pMIR125aΔ, a 58-bp segment of pMIR125a that encodes most of the pre-miR-125a stem-loop was deleted. Plasmid pMIR125b, also used to transfect 293T cells, was constructed from pCMV-Luc by replacing the luciferase translational unit with a 0.77-kb fragment of human chromosome 21 that encodes miR-125b flanked upstream by 484 nt and downstream by 270 nt. In plasmid pMIR125bΔ, a 59-bp segment of pMIR125b that encodes most of the pre-miR-125b stem-loop was deleted. Plasmid pMIR125b-U1A was derived from pMIR125b by introducing a T→A substitution at the position corresponding to the 5′-terminal nucleotide of miR-125b. Plasmid pSH-MIR125b, which was used to transfect P19 cells, was constructed by inserting a 0.08-kb gene fragment encoding pre-miR-125b downstream of the U6 promoter of pSHAG-1 (41). For use as a negative control, plasmid pCMV-con, which encodes no miRNA-related transcript whatsoever, was constructed from pCMV-Luc by deleting the luciferase translational unit.
The synthetic siDCR RNA duplex used to knock down Dicer synthesis was prepared by annealing complementary oligonucleotides that had been chemically synthesized (5′ AAAGGACCCAUUGGUGAGGAA 3′ and 5′ CCUCACCAAUGGGUCCUUUCU 3′; Dharmacon). The siGL2 RNA duplex used as a negative control comprised two complementary strands having the sequence 5′ CGUACGCGGAAUACUUCGAdTdT 3′ and 5′ UCGAAGUAUUCCGCGUACGdTdT 3′ (Dharmacon). The 2′-O-methyl oligonucleotide used to inhibit miR-125b function in transfected in P19 cells (UCACAAGUUAGGGUCUCAGGGA; Integrated DNA Technologies) and the nonspecific 2′-O-methyl oligonucleotide used as a negative control (AAGCGAAGCAGUGCGUCAAGUA) were purified by polyacrylamide gel electrophoresis before transfection.
Cell culture and transfection.
293T human embryonic kidney cells were grown in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10% fetal bovine serum. Transient transfection of 293T cells with DNA was performed for 12 h using GeneJuice (Novagen), following the manufacturer's protocol except that 1.5 ml of culture medium was used per 35-mm well. Transfected DNA mixtures (1 μg) contained a plasmid encoding a pri-miRNA (965 ng), a reporter plasmid (25 ng), and a plasmid encoding β-galactosidase (10 ng).
P19 mouse embryonal carcinoma cells were cultured and induced to differentiate into neurons essentially as described previously (16). In brief, undifferentiated P19 cells were grown in α-minimal essential medium (α-MEM) (GIBCO) supplemented with 10% fetal bovine serum. Differentiation of P19 cells was induced by treatment with retinoic acid (1 μM) for 4 days in 10-cm bacteriological petri dishes, which were coated with 0.2% agarose to prevent the cells from adhering. The cells were then disaggregated by treatment with trypsin, replated on poly-d-lysine-coated tissue culture dishes, and cultured in either α-MEM medium containing 10% fetal bovine serum or, in one instance, Neurobasal medium (GIBCO) containing B27 supplement (GIBCO) and glutamine (0.5 mM).
Transient transfection of undifferentiated P19 cells with DNA, siRNA, and/or 2′-O-methyl RNA was performed overnight by using Lipofectamine 2000 (3 to 5 μl; Invitrogen; see manufacturer's protocol) in tissue culture dishes with 12 23-mm wells. Transient transfection of differentiated P19 cells was carried out overnight using cells that had been treated with retinoic acid for 4 days. The cells were trypsinized, cultured for 24 h in tissue culture dishes with 12 23-mm wells, and transfected with DNA in the presence of Lipofectamine 2000 (2 to 3 μl). The transfection medium was replaced with fresh culture medium after 6 h, and cell extracts were prepared 30 h later.
Reporter assays.
Reporter lysis buffer (Promega) was used to prepare cell lysates for reporter expression assays. Luciferase activity was measured in a Tecan SpectraFluor Plus instrument by using the Bright-Glo luciferase assay system (Promega) according to the manufacturer's instructions. β-Galactosidase activity was assayed with o-nitrophenylgalactoside by spectrophotometry, as previously described (46), or with the Galacto-Light Plus chemiluminescent reporter assay system (Applied Biosystems). Each measurement of the ratio of luciferase to β-galactosidase activity was determined with lysates derived from at least three to four independent transfections that usually were performed on more than one day.
RNA assays.
For analysis of miRNA, total cellular RNA was extracted by using Trizol reagent (Invitrogen), according to the manufacturer's instructions. Equal amounts of total RNA (10 μg) were denatured, fractionated by electrophoresis on a 15% polyacrylamide-8 M urea gel, and electroblotted and cross-linked onto a Hybond-XL nylon membrane (Amersham). The blots were probed at 38°C in PerfectHyb Plus hybridization buffer (Sigma) with terminally radiolabeled DNA oligonucleotides complementary to miR-125a or miR-125b and then washed with 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) buffer at 38°C.
For luciferase, β-galactosidase, and lin-28 mRNA analysis, cytoplasmic RNA was purified from cells 36 h after transfection, as previously described (49), except that Trizol reagent (Invitrogen) was used to isolate RNA from cytoplasmic extracts. Equal amounts of cytoplasmic RNA (10 μg) were denatured by glyoxal treatment (46), fractionated by electrophoresis on a 1% agarose gel, and blotted and cross-linked onto a Hybond-XL nylon membrane (Amersham). The blots were probed at 68°C in PerfectHyb Plus hybridization buffer (Sigma) with radiolabeled luciferase DNA, β-galactosidase DNA, or lin-28 DNA prepared by random primer labeling using a High Prime DNA labeling kit (Roche) and then washed with 0.5× SSC-0.1% SDS buffer at 68°C.
Immunoblot assays.
Cells were harvested and lysed with RIPA lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing Complete protease inhibitor (Roche). Equal amounts of each lysate were separated by electrophoresis on a 6% polyacrylamide-SDS gel and electroblotted onto a Hybond ECL nitrocellulose membrane (Amersham). Each blot was blocked with 5% dry milk in phosphate-buffered saline containing 0.05% Tween 20, probed at 4°C overnight with antibodies against human Lin-28 (polyclonal rabbit antiserum, diluted 1:1,500) or actin (1:3000, Sigma), incubated with a secondary antibody conjugated to horseradish peroxidase (1:20,000; Bio-Rad), and developed with an Immun-Star HRP chemiluminescence kit (Bio-Rad).
RESULTS
miR-125-responsive elements in human lin-28 mRNA.
The nearly identical sequence of the 22-nucleotide human miRNA miR-125b suggests that it is the human homolog of C. elegans lin-4 (Fig. 1A) (23). A second human miRNA, miR-125a, differs from miR-125b at only four positions. The presence in humans of a gene encoding a protein homologous to the C. elegans lin-28 gene product raised the possibility that this human gene might be downregulated by miR-125a and/or miR-125b, much as the C. elegans lin-28 gene is repressed by lin-4. Consistent with this hypothesis, miR-125b levels rise and Lin-28 protein levels fall during mammalian embryonic development (21, 53; other data not shown). To determine whether either of these miRNAs regulates lin-28 gene expression in humans, we first developed a system for expressing miR-125a or miR-125b in a human cell line. Transient transfection of 293T human embryonic kidney cells with either of two human genes encoding primary RNA transcripts (pri-miR-125a or pri-miR-125b) that can be processed in vivo to produce miR-125a or miR-125b generated both the mature forms of these two miRNAs and their partially processed precursors (pre-miRNAs) at a significantly higher concentration than in untransfected cells (Fig. 1B). No such increase in abundance was observed for these two miRNAs in cells transfected with deletion mutants of the pri-miRNA genes from which the 58- to 59-bp segment encoding the immediate RNA precursor of miR-125a or miR-125b (the pre-miR-125a or pre-miR-125b stem-loop) had been removed.
FIG. 1.
Human miR-125a and miR-125b. A. Sequence of miR-125a, miR-125b, and C. elegans lin-4 miRNA. The miRNAs are drawn in a 5′-to-3′ orientation (left to right), and the conserved regions are highlighted. B. Increased abundance of miR-125a and miR-125b in transfected cells. 293T cells were transiently transfected either with a plasmid (pMIR125a or pMIR125b) encoding miR-125a or miR-125b or with a plasmid deletion variant (pMIR125aΔ or pMIR125bΔ) that did not encode an miRNA. After 36 h, total RNA was extracted and analyzed by gel electrophoresis and blotting, using radiolabeled oligonucleotide probes complementary to miR-125a or miR-125b. The sequence similarity of miR-125a and miR-125b resulted in some cross-hybridization of the two probes. miR-125a migrated as a doublet, suggesting two forms differing in length by one nucleotide, perhaps indicative of heterogeneity at the 3′-terminal processing site (±A). The sizes of the larger RNAs that were also detected suggest that they are partially processed stem-loop precursors of miR-125a and miR-125b (pre-miR-125a and pre-miR-125b). Unlike the miRNAs and pre-miRNAs in the cell extracts, the oligoribonucleotides used as approximate size markers (M) were not phosphorylated at the 5′ end and therefore migrated somewhat more slowly during electrophoresis than they would have otherwise.
To test whether the human lin-28 gene can be repressed by miR-125a and/or miR-125b, we constructed a reporter in which the 2.7-kb 3′ UTR of human lin-28 (Fig. 2A) was fused to a translational unit encoding firefly luciferase (Luc-lin 28). Expression of this reporter in transfected 293T cells was reduced by more than 80% upon cotransfection with a human gene encoding either miR-125a or miR-125b but not with otherwise identical miRNA genes lacking the pre-miR-125a or pre-miR-125b segment (Fig. 2B). These results indicate that the human lin-28 3′ UTR can function as a target for repression by both miR-125a and miR-125b.
FIG. 2.
Repression of luciferase reporters bearing lin-28 miREs by miR-125a and miR-125b. A. Map of human lin-28 mRNA and duplexes of the lin-28 miREs with miR-125a and miR-125b. In the RNA duplexes, the miRE (top strand) is drawn in a 5′-to-3′ orientation (left to right), and the miRNA (bottom strand) is drawn in a 3′-to-5′ orientation (left to right). These two lin-28 miREs are located directly beside one another, such that the two nucleotides shown at the 3′ end of miRE1 and at the 5′ end of miRE2 are the same two nucleotides of the lin-28 3′ UTR. B. Repression of a luciferase reporter bearing the lin-28 3′ UTR by miR-125a and miR-125b. 293T cells were transiently cotransfected with a luciferase reporter plasmid bearing either the human lin-28 3′ UTR (Luc-lin 28) or the SV40 late 3′ UTR (Luc), together with a plasmid encoding β-galactosidase (an internal standard to control for transfection efficiency) and one of five plasmids encoding either miR-125a or miR-125b, deletion variants thereof, or no miRNA-related transcript (control). Alternatively, the cotransfections were performed using Luc-lin 28 reporters from which miRE1 and/or miRE2 had been deleted. After 36 h, the ratio of luciferase activity to β-galactosidase activity in cell extracts was measured. Error bars correspond to the standard deviation of multiple measurements. C. Repression of a luciferase reporter bearing multiple copies of the lin-28 miREs by miR-125a and miR-125b. 293T cells were transiently cotransfected with a plasmid containing a luciferase-SV40 reporter gene (Luc) that bore 0, 2, 4, or 6 copies of miRE1, miRE2, or lin-28 element X (UGCAAGUGAGGGUUCUGGGGG) in its 3′ UTR and with one of four plasmids encoding miR-125a or miR-125b or deletion variants thereof, together with a plasmid encoding β-galactosidase (internal standard). After 36 h, the ratio of luciferase activity to β-galactosidase activity in cell extracts was measured. The activity ratio for cells transfected with genes encoding miR-125a or miR-125b was then normalized to the ratio in cells transfected with the corresponding deletion mutant that did not encode an miRNA and graphed.
Examination of the 3′ UTR of human lin-28 mRNA revealed countless segments with imperfect complementarity to these two miRNAs. The great abundance of these potential sites of regulation complicated the identification of the true regulatory elements. However, since human miR-125a and miR-125b are identical to their mouse counterparts, it seemed reasonable to assume that their miRE targets would also be conserved. By focusing on the 3′ UTR regions that are identical in human and mouse lin-28, we were able to identify two adjacent elements (miRE1 and miRE2) with the potential to base pair with miR-125a or miR-125b in a manner reminiscent of the imperfect duplexes predicted for lin-4 paired with its target elements in the C. elegans lin-14 and lin-28 mRNAs (9, 35, 52) (Fig. 2A). In both humans and mice, these two lin-28 elements are directly adjacent to one another. (On the basis of sequence examination, Moss and Tang [36] have also proposed that miRE2 might be a regulatory target of miR-125.) Deletion of both miRE1 and miRE2 from the 3′ UTR of the luciferase reporter nearly abolished repression by miR-125a and miR-125b, whereas deletion of either element alone reduced but did not eliminate repression (Fig. 2B). Whatever modest responsiveness remained in the reporter lacking both miRE1 and miRE2 was abolished when a third, weaker element (miRE0) (Fig. 3) was removed as well (because of its minimal efficacy, deleting miRE0 alone from the lin-28 3′ UTR had little influence on repression). Thus, miRE1 and miRE2 appear to be the principal elements within the human lin-28 3′ UTR that mediate repression by miR-125a and miR-125b, and both are necessary for efficient downregulation.
FIG. 3.
Nonresponsive or poorly responsive elements in the lin-28 3′ UTR. Hypothetical duplexes of nonresponsive or poorly responsive lin-28 3′ UTR elements (top strands, 5′-to-3′ orientation) with miR-125b (bottom strands, 3′-to-5′ orientation). The relative potency of each element in mediating translational repression by miR-125b in transfected 293T cells was calculated as (RE − 1)/(RmiRE1 − 1), where R is the ratio of luciferase production (normalized to β-galactosidase activity) in the absence versus the presence of miR-125b for a reporter bearing either two copies of the element (RE) or two copies of miRE1 (RmiRE1). Of the three elements shown, only miRE0 has detectable repression activity, and its effect is only 30% that of miRE1.
Additional experiments showed that miRE1 and miRE2 are alone sufficient to direct repression by miR-125a and miR-125b when each is inserted in multiple copies into another luciferase reporter bearing a 3′ UTR derived from SV40. The degree of repression was similar for miRE1 and miRE2 and was dependent on the number of miRE copies inserted (Fig. 2C) but not on their sequential order (1-2-1-2 versus 2-2-1-1 [data not shown]). By contrast, inserting multiple copies of other lin-28 3′ UTR elements with the potential to form thermodynamically favorable duplexes with miR-125a and miR-125b (element X or element Y) had no such effect, and insertion of miRE0 alone had only a small effect (Fig. 2C and 3). The repressive influence of miRE1 and miRE2 on reporter gene expression was also observed in mouse NIH 3T3 fibroblasts, which naturally produce miR-125b at a concentration similar to that in transfected 293T cells (data not shown).
Mutational analysis of miR-125-responsive elements.
That miRE1 is fairly effective at mediating the inhibitory effect of these miRNAs was somewhat surprising in view of previous reports that base pairing of the 5′-terminal segment of miRNAs with their target elements, especially miRNA nucleotides 2 to 8, is very important for repression, as is the overall free energy of duplex formation (3, 6, 20, 28). miRE1 can form consecutive Watson-Crick base pairs only with nucleotides 3 to 9 of miR-125a or miR-125b, unlike miRE2, which can base pair with nucleotides 1 to 9 (Fig. 2A). Furthermore, the duplex structures formed by miRE1 are expected to be of lower thermodynamic stability than those formed by miRE2. To investigate the basis for the efficacy of miRE1, we modified the 3′ portion of this element to alter its base-pairing potential with the 5′ segment of miR-125 and then tested the effect of these mutations on repression of a luciferase reporter containing two such elements (Fig. 4).
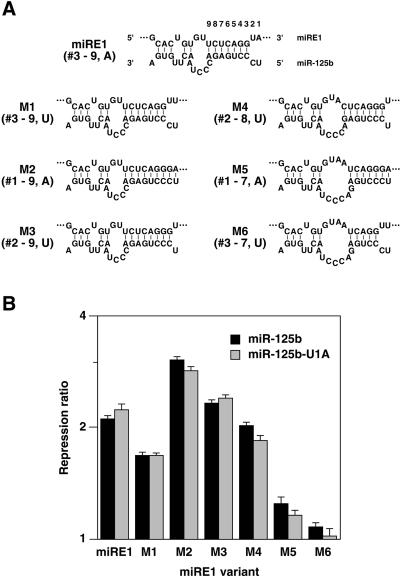
FIG. 4.
Efficacy of miRE1 variants in repressing gene expression. A. Duplexes of miRE1 and variants thereof (upper strands) with miR-125b (lower strands). Nucleotides in both strands of these duplexes are numbered from right to left. The miRE nucleotides expected to form consecutive base pairs with the 5′ portion of miR-125b are indicated beside each duplex, along with the identity of the nucleotide at the miRE 3′ end (position 1). The miRNA variant miR-125b-U1A (not shown) is identical to miR-125b except for a U→A substitution at the 5′ terminus (position 1). B. Repression of a luciferase-SV40 reporter gene bearing two copies of miRE1 or variants thereof. 293T cells were transiently cotransfected with plasmids encoding a reporter mRNA, β-galactosidase (internal standard), and either miR-125b, an miR-125b substitution mutant bearing a 5′terminal adenosine (miR-125b-U1A), or an miR-125b deletion mutant (miR-125bΔ). After 36 h, the ratio of luciferase activity to β-galactosidase activity in cell extracts was measured. By dividing the ratio in cells lacking miR-125b by the ratio in otherwise identical cells containing miR-125b (black bars) or miR-125b-U1A (gray bars), repression ratios were calculated for each 3′ UTR element. These repression ratios were then graphed on a logarithmic scale.
Our data indicate that the presence of an adenosine at the 3′ end of miRE1 (position 1) is important for the function of this RNA element. Without this adenosine residue (M1), miRE1 became significantly less active. The contribution of the adenosine at position 1 was also evident for an miRE1 variant (M2) whose efficacy had been increased by changing the adjacent nucleotide to allow base pairing at position 2; in this context as well, an A→U substitution at position 1 (M3) was deleterious. Among the miRE1 variants that could form seven consecutive base pairs with the 5′ segment of miR-125b, pairing with nucleotides 2 to 8 (M4) or 3 to 9 (M1) was more effective than pairing with nucleotides 1 to 7 (M5).
Additional experiments showed that the contribution of the 3′-terminal adenosine residue to miRE1 function is not a consequence of its complementarity to the uridine at position 1 of miR-125b. This was determined by changing the first nucleotide of miR-125b from U to A (miR-125b-U1A) and measuring the efficacy of the modified miRNA in downregulating the same set of reporters, each of which contains a pair of miREs that end with either an adenosine or a uridine residue. To within experimental error, the mutant and wild-type forms of miR-125b were indistinguishable in their ability to repress every sequence variant of miRE1, regardless of the base-pairing potential of the 3′-terminal miRE residue (Fig. 4B). That an adenosine at this position can enhance miRE1 function irrespective of its ability to base pair may also help to explain the lesser activity of miRE0 and the inactivity of element Y, both of which can pair with nucleotides 3 to 9 of miR-125b but lack a terminal adenosine residue opposite nucleotide 1 (Fig. 3).
Influence of miR-125 on mRNA abundance.
Gene regulation by miRNAs that are imperfectly complementary to their mRNA targets is generally thought to be achieved via a translational repression mechanism (5, 38, 40, 52, 56), whereas silencing by perfectly complementary siRNAs typically involves accelerated mRNA degradation (33, 51). To determine whether repression mediated by miRE1 and miRE2 occurs only at the level of translation, the cytoplasmic concentration of luciferase reporter mRNAs bearing six copies of miRE1 or miRE2 was compared in the presence or absence of miR-125a or miR-125b. The significant decline in luciferase protein synthesis observed in cells that produced either of these miRNAs was accompanied by a smaller but significant decrease in the concentration of the luciferase reporter mRNA (Fig. 5). In each case (and in the case of a reporter containing the entire lin-28 3′ UTR; data not shown), the calculated reductions in mRNA abundance and translational efficiency (protein yield per mRNA molecule) were approximately equal in magnitude. No such change in either mRNA or protein concentration was observed for a luciferase control gene that lacked the lin-28 miREs but was expressed under the control of the same promoter.
FIG. 5.
Relative contributions of translational efficiency and mRNA abundance to repression by miR-125a and miR-125b. A. RNA blot showing the effect of miR-125a and miR-125b on the cellular concentration of a reporter mRNA containing zero or six copies of lin-28 miRE1 or miRE2. 293T cells were transiently cotransfected with a reporter gene, a β-galactosidase gene, and a gene encoding miR-125a (a), miR-125b (b), or an otherwise identical primary transcript from which the pre-miRNA stem-loop had been deleted (Δ). After 36 h, cytoplasmic RNA was isolated and analyzed by gel electrophoresis and blotting, using radiolabeled probes complementary to luciferase mRNA and β-galactosidase mRNA (internal standard). B. Contributions of translational inhibition and diminished mRNA abundance to repression by miR-125a and miR-125b. The reporter mRNA concentration in each sample was normalized to the concentration of β-galactosidase mRNA, and the ratio of the normalized concentration of the reporter mRNA in cells transfected with an miR-125a or miR-125b gene versus the corresponding miRNA deletion variant was calculated. To reveal the relative contributions of translational efficiency (protein yield per mRNA molecule) and mRNA abundance to repressing gene expression, the calculated effect of each miRNA on the cytoplasmic abundance of reporter mRNAs bearing six copies of miRE1 or miRE2 was superposed on a graph showing the overall degree of repression of the same reporter genes by each miRNA. This overall degree of repression, the repression ratio, was calculated as for Fig. 4. The effect of an miRNA on translational efficiency (black bars) corresponds to the ratio of its overall effect on protein synthesis versus its effect on mRNA abundance (gray bars). In the case of miRE1, expression of miR-125a or miR-125b reduced translational efficiency by a factor of 4.3 ± 0.3 or 2.9 ± 0.4, respectively, and mRNA abundance by a factor of 4.2 ± 0.2 or 3.2 ± 0.1, respectively. In the case of miRE2, expression of miR-125a or miR-125b reduced translational efficiency by a factor of 3.8 ± 0.4 or 2.2 ± 0.3, respectively, and mRNA abundance by a factor of 3.1 ± 0.3 or 2.7 ± 0.3, respectively.
In mammalian cells, siRNAs can reduce the concentration of targeted mRNAs not only by base pairing with perfectly complementary RNA elements to direct mRNA cleavage but also by acting on complementary DNA elements to cause transcriptional silencing (17, 34). If the miRE-dependent reduction in mRNA concentration that is mediated by miR-125a and miR-125b occurs via a mechanism unrelated to transcription initiation, then this reduction should be promoter independent (as is observed; see below) and should require that the miREs be present in the mRNA and not merely encoded in the DNA. To address the latter point, a 0.23-kb DNA fragment encoding an intact or mutated polyadenylation signal (AAUAAA or UUCUUU) was inserted into a luciferase reporter gene between the coding region and six copies of miRE1 present in the 3′ UTR. The two resulting reporters, which were almost identical in DNA sequence but encoded mRNA transcripts that either contained or lacked the miREs depending on the site of polyadenylation, were tested in 293T cells for their responsiveness to miR-125b (Fig. 6). Only the reporter that bore the defective polyadenylation signal and therefore encoded a longer mRNA containing the miREs was repressed, as evidenced by an miR-125b-dependent reduction in both luciferase protein synthesis and reporter mRNA abundance. In contrast, the presence of an efficient polyadenylation site upstream of the miREs abolished repression and rendered the reporter mRNA concentration insensitive to miR-125b. Together, these findings suggest that the interaction of miR-125a and miR-125b with miRE1 and miRE2 of human lin-28 represses gene expression by a dual mechanism involving both an impediment to translation and a posttranscriptional reduction in mRNA abundance.
FIG. 6.
Consequences of inserting a polyadenylation site upstream of the miREs. A. Luciferase reporter gene bearing an intact (wt: AATAAA) or defective (mut: TTCTTT) bovine growth hormone (BGH) polyadenylation signal upstream of six copies of lin-28 miRE1 and an intact SV40 polyadenylation signal downstream of the miREs. B. Effect of miR-125b on the expression of reporter genes in which an intact (wt) or defective (mut) BGH polyadenylation signal had been inserted into the 3′ UTR upstream of zero or six copies of lin-28 miRE1. 293T cells were transiently cotransfected with a reporter gene, a β-galactosidase gene, and a gene encoding miR-125b or an otherwise identical primary transcript from which the pre-miRNA stem-loop had been deleted (miR-125bΔ). After 36 h, the ratio of luciferase activity to β-galactosidase activity in cell extracts was measured. C. RNA blot showing the effect of miR-125b on the cytoplasmic concentration of a reporter mRNA in which an intact (wt) or defective (mut) BGH polyadenylation signal had been inserted upstream of six copies of lin-28 miRE1. 293T cells were transiently cotransfected as in A, and after 36 h, cytoplasmic RNA was isolated and analyzed as for Fig. 5. Compared to a negative control (Δ = miR-125bΔ), miR-125b (b) reduced the cytoplasmic concentration of the 2.4-kb reporter mRNA bearing a defective polyadenylation signal and six copies of miRE1 to 44% ± 4% of its unrepressed value but did not affect the concentration of the 1.8-kb mRNA transcribed from an otherwise identical gene bearing a functional polyadenylation signal upstream of the miREs (98% ± 7%).
Further investigation revealed that the observed reduction in mRNA concentration was not a secondary consequence of the decrease in translational efficiency caused by interaction of miR-125 with the lin-28 miREs. A large (42-nucleotide) stem-loop structure was inserted into the 5′ UTR of a luciferase reporter transcript containing six copies of miRE1 in its 3′ UTR. By blocking ribosome scanning, the stem-loop alone virtually abolished translation (99% inhibition of luciferase synthesis), yet its presence had no effect on either the abundance of the reporter mRNA in the absence of miR-125b or the ability of this miRNA to cause a marked reduction in the cellular concentration of the reporter message (Fig. 7). Thus, impaired translation per se is not sufficient to cause the concentration of this mRNA to fall, nor is translation required for miR-125b to mediate a decline in reporter mRNA abundance. We conclude that miR-125b reduces the concentration of mRNA containing the lin-28 miREs by a posttranscriptional mechanism that is independent of its inhibitory effect on translation.
FIG. 7.
Translation independence of the downregulation of mRNA abundance by miR-125b. 293T cells were transiently cotransfected with a luciferase reporter gene bearing six copies of miRE1 in the 3′ UTR, with or without a segment encoding a large stem-loop structure (CGGGGCGCGUGGUGGCGGCUGCAGCCGCCACCACGCGCCCCG) in the 5′ UTR, a β-galactosidase gene, and a gene encoding miR-125b (b) or an otherwise identical primary transcript from which the pre-miRNA stem-loop had been deleted (Δ). After 36 h, cytoplasmic RNA was isolated and analyzed as for Fig. 5. The ratio of the reporter mRNA to β-galactosidase mRNA (internal standard) is indicated at the bottom of each lane.
Role of miR-125 in downregulating lin-28 during neuronal differentiation.
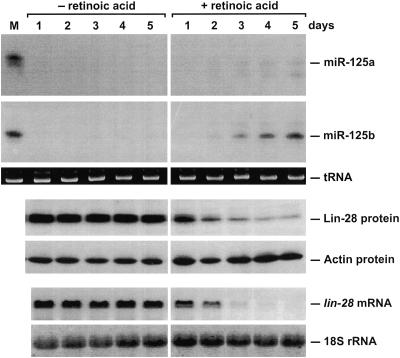
Although 293T cells can be genetically engineered for use in studies of repression by miRNAs, this cell line does not normally produce lin-28 mRNA, miR-125a, or miR-125b. To examine the role of miR-125a and miR-125b in regulating expression of the endogenous lin-28 gene in mammalian cells, we instead chose P19 embryonal carcinoma cells. This mouse cell line has been widely used as a model system for investigating the differentiation of nerve cells in mammals, since it can be induced to differentiate into neurons, astrocytes, and fibroblast-like cells upon treatment with retinoic acid (16); in addition, it is known to express the lin-28 gene (36). Prior to induction of P19 cells, miR-125a and miR-125b are virtually undetectable, whereas the mRNA and protein products of the endogenous lin-28 gene are quite abundant (Fig. 8). Over a period of 4 to 5 days following induction with retinoic acid, the cellular concentration of miR-125b and, to a lesser extent, miR-125a increases significantly, while Lin-28 protein and mRNA levels decline markedly (Fig. 8) (48, 53). (The abundance of miR-125b in P19 cells treated with retinoic acid for 7 days is even higher than that in 293T cells transfected with a gene encoding this miRNA.) Thus, lin-28 gene expression in differentiating P19 cells is inversely correlated with the cellular concentrations of miR-125a and miR-125b, a finding consistent with a natural role for these miRNAs in repressing lin-28 expression.
FIG. 8.
Changes in miR-125a and miR-125b levels and lin-28 gene expression following retinoic acid induction of P19 cell differentiation. P19 cells were grown for 5 days in agarose-coated petri dishes in the presence or absence of retinoic acid (1 μM). RNA and protein samples were extracted daily, and equal amounts were analyzed by blotting, using radiolabeled DNA probes to detect specific RNAs (miR-125a, miR-125b, and lin-28 mRNA) and polyclonal anti-Lin-28 antibodies to detect Lin-28. tRNA, 18S rRNA, and actin served as internal standards. Lane M, RNA extracted from 293T cells transiently transfected with either pMIR125a (top pair of panels; detection with an miR-125a-specific probe) or pMIR125b (second pair of panels; detection with an miR-125b-specific probe); note the similar abundance of miR-125b in P19 cells induced for 5 days and in transfected 293T cells. Due to low-level cross-hybridization, miR-125b (faint lower band) was detectable with the miR-125a-specific probe, and miR-125a (faint upper band) was detectable with the miR-125b-specific probe.
To test the validity of this inference, we impeded miRNA production in P19 cells by using RNA interference to knock down expression of Dicer, the RNase responsible for the last step in miRNA processing. Transfection of retinoic acid-treated P19 cells with Dicer siRNA reduced miR-125b levels by 72% and increased the cellular abundance of the Lin-28 protein by almost a factor of two versus cells transfected with a control siRNA (data not shown). We conclude that the repression of lin-28 following induction of P19 cells is, at least in part, miRNA dependent.
That miR-125b in particular likely contributes to lin-28 repression during differentiation of P19 cells was implied by experiments showing that this miRNA is able to downregulate expression of the endogenous lin-28 gene in undifferentiated P19 cells engineered to produce miR-125b at a physiological concentration. Uninduced P19 cells were transfected with a plasmid encoding pre-miR-125b so as to raise the cellular abundance of the corresponding miRNA to a level comparable to that in retinoic acid-treated cells (Fig. 9) and in rat hippocampal neurons 3 days before birth (data not shown). The increased concentration of miR-125b in these cells resulted in a marked reduction in Lin-28 protein synthesis compared to that in P19 cells transfected with a control plasmid that did not encode the miRNA (Fig. 9). This reduction in protein synthesis was accompanied by a significant decrease in the cytoplasmic concentration of lin-28 mRNA, as observed in 293T cells for reporter mRNAs bearing the lin-28 miREs but transcribed under the control of a different promoter. These findings indicate that even in the absence of differentiation, miR-125b is alone sufficient to cause significant repression of lin-28 gene expression in P19 cells, a conclusion consistent with a similar role for this miRNA in differentiating P19 cells.
FIG. 9.
Downregulation of lin-28 gene expression by production of miR-125b in undifferentiated P19 cells. Undifferentiated P19 cells were transfected with a plasmid (pSH-MIR125b, 1.5 μg) encoding miR-125b or with a variant thereof (pSHAG-1, 1.5 μg) lacking the miR-125b gene segment (Δ), and after two days, RNA and protein extracts were prepared. Alternatively, untransfected P19 cells (−) were treated with retinoic acid for 4 days, and RNA was extracted 1 day later. Equal amounts of each RNA or protein sample were analyzed by blotting to detect miR-125b, Lin-28 protein, and lin-28 mRNA. tRNA, 18S rRNA, and actin served as internal standards. The increased production of miR-125b in undifferentiated P19 cells that had been transfected with pSH-MIR125b reduced the abundance of the Lin-28 protein and lin-28 mRNA to approximately one-quarter (26% ± 2%) and one-half (52% ± 2%) of their respective concentrations in control cells (Δ).
To demonstrate directly that miR-125b helps to downregulate lin-28 gene expression in P19 cells that are undergoing differentiation, we selectively inhibited the function of this miRNA by transfection with a complementary 2′-O-methyl oligonucleotide (13, 32). In uninduced P19 cells, this complementary oligonucleotide interfered with the capacity of plasmid-encoded miR-125b to repress the expression of a cotransfected luciferase reporter bearing six copies of miRE2 (Fig. 10A). In P19 cells induced to differentiate, the presence of the complementary oligonucleotide impaired the ability of endogenous miR-125b to repress Lin-28 protein synthesis (Fig. 10B).
FIG. 10.
Influence of miR-125b on Lin-28 protein synthesis in differentiating P19 cells. A. (Top) Sequence of miR-125b and of the complementary 2′-O-methylated oligonucleotide (2′ OMe anti-miR-125b) used to repress its activity. Also shown is the sequence of a noncomplementary 2′-O-methylated oligonucleotide used as a negative control. (Bottom) The efficacy of the 2′ OMe anti-miR-125b oligonucleotide in repressing miR-125b function was confirmed by transiently cotransfecting undifferentiated P19 cells with a luciferase-SV40 reporter gene bearing zero or six copies of miRE2 in its 3′ UTR (25 ng), a plasmid encoding miR-125b or a deletion variant thereof (750 ng), a plasmid encoding β-galactosidase (25 ng, internal standard), and either the 2′ OMe anti-miR-125b oligonucleotide or the control oligonucleotide (100 pmol). After 48 h, cell extracts were prepared, and the ratio of luciferase activity to β-galactosidase activity was measured. The activity ratio in cells transfected with a reporter containing six copies of miRE2 was then normalized to the ratio in cells transfected with the corresponding reporter lacking this element, and the ratios were graphed. B. Undifferentiated P19 cells were transiently transfected with the 2′ OMe anti-miR-125b oligonucleotide or the negative control 2′ OMe oligonucleotide (200 pmol) and then induced to differentiate by treatment with retinoic acid. Five-and-a-half days after induction, protein extracts were prepared, and equal amounts of each extract were analyzed by immunoblotting to detect Lin-28 protein and actin (an internal standard). Compared to the control oligonucleotide, transfection with the 2′-O-methylated anti-miR-125b oligonucleotide increased Lin-28 protein synthesis by almost a factor of 2 (1.8 ± 0.3).
To confirm that the miR-125-responsive elements in the lin-28 gene contribute to its downregulation in differentiating P19 cells, we compared the expression of a luciferase reporter gene bearing the entire lin-28 3′ UTR (Luc-lin 28) to that of an otherwise identical reporter from which these elements (miRE0, miRE1, and miRE2) had been deleted. In P19 cells treated with retinoic acid to induce miR-125 production, expression of the reporter bearing an intact lin-28 3′ UTR was about half that of the reporter lacking the three miREs (Fig. 11A). By contrast, little if any decrease in Luc-lin 28 expression was observed in uninduced cells devoid of miR-125 (Fig. 11A). Similarly, compared to a reporter that lacked miREs, the expression of luciferase reporters containing six copies of miRE1 or miRE2 was reduced by 60% in retinoic acid-treated cells and was diminished to an even greater extent when the induced P19 cells were cultured in a medium (Neurobasal/B27 [4]) that favored neuronal cells over other differentiated cell fates (Fig. 11B; other data not shown). These results imply that the miR-125-responsive elements in lin-28 help to downregulate this gene in P19 cells undergoing differentiation into neurons.
FIG. 11.
Contribution of lin-28 miREs to repressing gene expression in differentiating P19 cells. A. P19 cells that had (+) or had not (−) been treated with retinoic acid were transiently cotransfected with a plasmid encoding a luciferase reporter fused to the human lin-28 3′ UTR (Luc-lin 28, 0.05 μg), a plasmid encoding β-galactosidase (0.05 μg, an internal standard), and pUC19 (0.9 μg). Alternatively, the cotransfection was performed using Luc-lin 28 reporters from which elements responsive to miR-125 (miRE0, miRE1, and miRE2) and/or let-7 miRNA (L7: GAGUGCACAGCCUAUUGAACUACCUCA) had been deleted (see map of human lin-28 mRNA above graph). After 36 h, cell extracts were prepared, and the ratio of luciferase activity to β-galactosidase activity was measured. Prior to graphing, the ratios were normalized to the values measured for the reporter lacking all four miREs. B. P19 cells were transiently cotransfected with a plasmid encoding a luciferase-SV40 reporter mRNA that bore zero or six copies of miRE1 in its 3′ UTR (0.7 μg) and a plasmid encoding β-galactosidase (0.9 μg, internal standard). After 12 h, the cells were transferred to petri dishes and cultured for 4 days in α-MEM containing fetal bovine serum (FBS) (10%) and retinoic acid (1 μM). The cells were then transferred to tissue culture dishes coated with poly-d-lysine and cultured for six more days in either α-MEM containing 10% fetal bovine serum (to allow the growth of nonneuronal cells) or Neurobasal medium containing B27 supplement (to suppress the growth of nonneuronal cells) (4). Cell extracts were prepared at 2-day intervals, and the ratio of luciferase activity to β-galactosidase activity was measured. The activity ratio in cells transfected with the reporter that contained six copies of miRE1 was then normalized to the ratio in cells transfected with the reporter that lacked such miREs, and the data were graphed.
Additional segments within the lin-28 3′ UTR bear significant complementarity to other miRNAs that increase in abundance when P19 cells are treated with retinoic acid (let-7, miR-9, miR-30, and miR-128), raising the possibility that lin-28 may be repressed by multiple miRNAs (14, 27, 36, 38, 48). However, among these potential regulatory elements, only deletion of the segment complementary to let-7 (L7) had any influence on expression of the Luc-lin 28 reporter in retinoic acid-treated cells, increasing luciferase synthesis by ∼40% relative to that of a reporter with an intact 3′ UTR (Fig. 11A). The regulatory effects of this element and the miR-125-responsive elements within the lin-28 3′ UTR were additive. We conclude that let-7 makes a smaller, but significant, contribution to lin-28 repression in differentiating P19 cells.
DISCUSSION
Our findings indicate that the increased cellular abundance of miR-125b that accompanies neuronal differentiation of retinoic acid-treated P19 cells makes a significant contribution to the concomitant repression of lin-28 in these cells. miR-125b inhibits Lin-28 protein synthesis by interacting with two strong elements (miRE1 and miRE2) and one weak element (miRE0) in the lin-28 3′ UTR. The repressive effect of these interactions, while smaller in magnitude than the overall degree to which lin-28 is downregulated upon differentiation of P19 cells, is of the size expected for a gene that contains only two to three elements that are responsive to a particular miRNA and appears to be augmented by the repressive effect of at least one other miRNA (let-7); further repression by a mechanism operating at the level of transcription is also possible (26, 48). By extrapolation, miR-125b may play a significant role in mammalian nervous system development by contributing to the repression of lin-28 (and possibly other genes) during neuronal differentiation. Consistent with this view are the rising concentration of miR-125b and the falling concentration of the Lin-28 protein in the developing brains of rodents (21, 53) and the reduced expression of the neuronal marker β-tubulin III in differentiating P19 cells in which gene regulation by miR-125b has been impaired (data not shown). Thus, contrary to a recent conclusion from studies of Tera-2 cells (26), it appears that the ability of lin-4-like miRNAs to regulate lin-28 gene expression via miREs in the lin-28 3′ UTR is conserved from nematodes to humans.
The efficacy of miRE1 and miRE2 as regulatory targets of miR-125b and its homolog miR-125a is not merely a consequence of their potential for stable base pairing with these miRNAs. These two lin-28 elements are similarly potent in mediating repression despite significant differences in the length and thermodynamic stability of the duplex that each can form with the 5′ segment of miR-125a and miR-125b, the miRNA segment believed to be most important for determining miRE activity (6, 19, 20, 28). Whereas miRE2 can base pair with the first nine nucleotides of miR-125, a property consistent with previous reports as to the importance of base pairing to miRNA nucleotides 2 to 8, miRE1 can form consecutive Watson-Crick base pairs only with nucleotides 3 to 9. Our data indicate that the presence in miRE1 of an adenosine opposite the 5′-terminal nucleotide of miR-125 (nucleotide 1) enables this element to function well regardless of the base-pairing potential of this adenosine residue and despite the inability of the adjacent miRE residue to pair with the second nucleotide of miR-125. By extrapolation, this finding offers an explanation for the preponderance of adenosines at this position in predicted vertebrate miREs irrespective of the identity of the 5′-terminal miRNA nucleotide (27), and it suggests that the protein complex surrounding the miRE-miRNA duplex may have a surface for binding this adenosine nucleotide. These results may also help to explain why two other lin-28 elements that can form thermodynamically favorable duplexes with miR-125 (miRE0 and element Y) do not function well as miREs, since neither has a 3′-terminal adenosine or is capable of base pairing with the second nucleotide of miR-125. On the other hand, the inactivity of element X as an miR125a/b target is likely due to the multiple G-U base pairs that it would form with the 5′ portion of these two miRNAs, since wobble pairs in this crucial region are thought to be detrimental to miRNA function (6).
Initial studies with mammalian cells of genetic repression mediated by artificial or natural miREs partially complementary to an miRNA found that diminished reporter protein synthesis was not accompanied by a significant reduction in mRNA abundance (5, 38, 56). These and similar results from earlier studies of miRNA function in C. elegans (40, 47, 52) led to the conclusion that unlike the degradative repression mechanism of siRNAs (and at least one mammalian miRNA [54]) that are perfectly complementary to their mRNA targets, repression by imperfectly complementary miRNAs might occur exclusively at the level of translation. However, we have discovered that miR-125b causes not only a reduction in Lin-28 protein levels but also a substantial decrease in lin-28 mRNA abundance that is dependent on the presence of the miREs within the message but independent of both the mRNA context of the miREs and the promoter that controls mRNA synthesis. We have also found that in mammalian cells, let-7 causes a similar reduction in the concentration of a reporter mRNA containing multiple copies of the lin-28 L7 element (data not shown). These results suggest that the means by which miR-125b and let-7 repress lin-28 expression in mammals involves both impaired translation and a posttranscriptional mechanism for reducing the mRNA concentration, possibly via accelerated mRNA degradation. Consistent with these observations is a recent report that transfection of human cells with miR-1 or miR-124 can diminish the abundance of certain mRNAs (30), although those experiments did not establish whether the effect was posttranscriptional or whether the mRNA elements responsible for repressing gene expression were also necessary and sufficient to cause a concomitant change in mRNA concentration. Furthermore, our data indicate that the reduction in mRNA abundance mediated by miR-125b is not a secondary consequence of its inhibitory effect on translation. Whatever the mechanism may be, these findings indicate that the regulatory processes used by miRNAs to repress mRNAs to which they are imperfectly complementary or by siRNAs to silence messages with perfect complementarity may in some instances be less distinct than originally thought.
Acknowledgments
We thank Zhai Li for advice on tissue culture.
These studies were supported by a research grant to J.G.B. from the National Institutes of Health (GM55624).
REFERENCES
- 1.Ambros, V., and H. R. Horvitz. 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226:409-416. [DOI] [PubMed] [Google Scholar]
- 2.Berezikov, E., V. Guryev, J. van de Belt, E. Wienholds, R. H. Plasterk, and E. Cuppen. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120:21-24. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke, J., A. Stark, R. B. Russell, and S. M. Cohen. 2005. Principles of microRNA-target recognition. PloS. Biology 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewer, G. J., J. R. Torricelli, E. K. Evege, and P. J. Price. 1993. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35:567-576. [DOI] [PubMed] [Google Scholar]
- 5.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doench, J. G., and P. A. Sharp. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18:504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 9.Ha, I., B. Wightman, and G. Ruvkun. 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 10:3041-3050. [DOI] [PubMed] [Google Scholar]
- 10.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 11.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 12.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 13.Hutvagner, G., M. J. Simard, C. C. Mello, and P. D. Zamore. 2004. Sequence-specific inhibition of small RNA function. PloS Biol. 2:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human MicroRNA targets. PloS Biol. 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, S. M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown, and F. J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120:635-647. [DOI] [PubMed] [Google Scholar]
- 16.Jones-Villeneuve, E. M., M. W. McBurney, K. A. Rogers, and V. I. Kalnins. 1982. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J. Cell Biol. 94:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasaki, H., and K. Taira. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431:211-217. [DOI] [PubMed] [Google Scholar]
- 18.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiriakidou, M., P. T. Nelson, A. Kouranov, P. Fitziev, C. Bouyioukos, Z. Mourelatos, and A. Hatzigeorgiou. 2004. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 18:1165-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloosterman, W. P., E. Wienholds, R. F. Ketting, and R. H. Plasterk. 2004. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 32:6284-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krichevsky, A. M., K. S. King, C. P. Donahue, K. Khrapko, and K. S. Kosik. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9:1274-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagos-Quintana, M., R. Rauhut, W. Lendeckel, and T. Tuschl. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853-858. [DOI] [PubMed] [Google Scholar]
- 23.Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735-739. [DOI] [PubMed] [Google Scholar]
- 24.Lee, R. C., R. L. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843-854. [DOI] [PubMed] [Google Scholar]
- 25.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 26.Lee, Y. S., H. K. Kim, S. Chung, K. S. Kim, and A. Dutta. 2005. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J. Biol. Chem. 280:16635-16641. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15-20. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115:787-798. [DOI] [PubMed] [Google Scholar]
- 29.Lim, L. P., M. E. Glasner, S. Yekta, C. B. Burge, and D. P. Bartel. 2003. Vertebrate microRNA genes. Science 299:1540. [DOI] [PubMed] [Google Scholar]
- 30.Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, and J. M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769-773. [DOI] [PubMed] [Google Scholar]
- 31.Lin, S. Y., S. M. Johnson, M. Abraham, M. C. Vella, A. Pasquinelli, C. Gamberi, E. Gottlieb, and F. J. Slack. 2003. The C. elegans hunchback Homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4:639-650. [DOI] [PubMed] [Google Scholar]
- 32.Meister, G., M. Landthaler, Y. Dorsett, and T. Tuschl. 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montgomery, M. K., S. Xu, and A. Fire. 1998. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95:15502-15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris, K. V., S. W. Chan, S. E. Jacobsen, and D. J. Looney. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305:1289-1292. [DOI] [PubMed] [Google Scholar]
- 35.Moss, E. G., R. C. Lee, and V. Ambros. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88:637-646. [DOI] [PubMed] [Google Scholar]
- 36.Moss, E. G., and L. Tang. 2003. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 258:432-442. [DOI] [PubMed] [Google Scholar]
- 37.Mourelatos, Z., J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16:720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, P. T., A. G. Hatzigeorgiou, and Z. Mourelatos. 2004. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 10:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839-843. [DOI] [PubMed] [Google Scholar]
- 40.Olsen, P. H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671-680. [DOI] [PubMed] [Google Scholar]
- 41.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasquinelli, A. E., B. J. Reinhart, F. Slack, M. Q. Martindale, M. I. Kuroda, B. Maller, D. C. Hayward, E. E. Ball, B. Degnan, P. Muller, J. Spring, A. Srinivasan, M. Fishman, J. Finnerty, J. Corbo, M. Levine, P. Leahy, E. Davidson, and G. Ruvkun. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408:86-89. [DOI] [PubMed] [Google Scholar]
- 43.Poy, M. N., L. Eliasson, J. Krutzfeldt, S. Kuwajima, X. Ma, P. E. Macdonald, S. Pfeffer, T. Tuschl, N. Rajewsky, P. Rorsman, and M. Stoffel. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432:226-230. [DOI] [PubMed] [Google Scholar]
- 44.Reinhart, B. J., F. J. Slack, M. Basson, A. E. Pasquinelli, J. C. Bettinger, A. E. Rougvie, H. R. Horvitz, and G. Ruvkun. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901-906. [DOI] [PubMed] [Google Scholar]
- 45.Reinhart, B. J., E. G. Weinstein, M. W. Rhoades, B. Bartel, and D. P. Bartel. 2002. MicroRNAs in plants. Genes Dev. 16:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Seggerson, K., L. Tang, and E. G. Moss. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243:215-225. [DOI] [PubMed] [Google Scholar]
- 48.Sempere, L. F., S. Freemantle, I. Pitha-Rowe, E. Moss, E. Dmitrovsky, and V. Ambros. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shyu, A.-B., M. E. Greenberg, and J. G. Belasco. 1989. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3:60-72. [DOI] [PubMed] [Google Scholar]
- 50.Smalheiser, N. R., and V. I. Torvik. 2004. A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions. BMC Bioinformatics 5:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuschl, T., P. D. Zamore, R. Lehmann, D. P. Bartel, and P. A. Sharp. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13:3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wightman, B., I. Ha, and G. Ruvkun. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855-862. [DOI] [PubMed] [Google Scholar]
- 53.Yang, D. H., and E. G. Moss. 2003. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr. Patterns 3:719-726. [DOI] [PubMed] [Google Scholar]
- 54.Yekta, S., I. H. Shih, and D. P. Bartel. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304:594-596. [DOI] [PubMed] [Google Scholar]
- 55.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 56.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed microRNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436:214-220. [DOI] [PubMed] [Google Scholar]